Identifying the Right Samples: A Guide to Metabolomics Biomarker Research
Metabolomics Biomarker Series Article Catalog:
1. Unlocking Biomarkers: A Guide to Vital Health Indicators
2. Metabolomics and Biomarkers: Unveiling the Secrets of Biological Signatures
3. Choosing the Right Study Design for Metabolomics Biomarker Discover
4. Metabolomics Biomarker Screening Process
All excellent scientific research results are based on a good "protocol design". It is, therefore, extremely important to develop a research protocol for metabolic marker screening. The steps in conducting a metabolomics clinical marker study including determining study type, designing screening process, calculating sample size and developing sample criteria. In previous sections, we have known how to select the most suitable study type and design the screening process. Then, the calculation of the sample size is required for each phase. Clinical samples constitute the basis for biomarker screening. Therefore, when addressing the clinical question, it is important to consider the volume of samples that can be collected - adequate samples must be available from the initial screening to the later validation phase. There are various sample sources, with the basic principle being that samples that can be obtained non-invasively or minimally invasively are preferred. Serum, plasma, urine, saliva, and even cerebrospinal fluid can be used as screening samples. However, metabolomics results are susceptible to external influences (such as the environment). Therefore, strict inclusion criteria for samples must be applied to screen for true-positive metabolic biomarkers. Moreover, clinical information, including patient's clinical data, pathologic diagnosis, follow-up records, etc., is also important - such information underlies the later development of the diagnostic panel and contributes significantly to the reliability of the biomarker. In this chapter, we will discuss how to estimate the specific sample size and set the sample standards.
1. Calculating sample size
Important parameters affecting sample size calculation are as follows:
① Type of study design (e.g., cross-sectional study or randomized controlled study)
② Type of outcome indicator (dichotomous variable or continuous variable)
③ Expected value of the outcome indicator (sensitivity /specificity /mean /AUC)
④ Allowable error
⑤ Test level α (usually taken as α=0.05)
⑥ Test efficacy 1-β (usually 80% or higher)
⑦ Ratio of case group size/control group size ratio: It is generally not advisable to have a smaller sample size in the control group than in the case group, typically of equal size.
Next, we will outline how sample size is calculated for a single-sample clinical diagnostic experiment for a cross-sectional study.
Calculation based on sensitivity and specificity
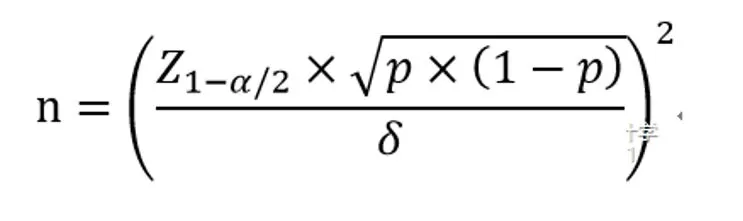
n represents the sample size for each group; α=0.05; when α is taken as 0.01, Z1α/2 = 2.58.
The Z1-α/2 value needs to be looked up in the table. In this case, Z1-α/2 =1.96.
δ represents the allowable error.
p stands for sensitivity (Pse) or specificity (Psp).
δ refers to the width of the 95% interval of the allowable sensitivity or specificity, which is assigned artificially by the investigator and is usually set at 0.03 to 0.1.
For example, if we set the parameters to:
Specificity: 90% ± 10%;
Sensitivity: 80% ± 10%;
Significance level (α): 0.05;
Confidence level (1- α): 0.95;
Test type: two-sided test
Calculation based on the formula:
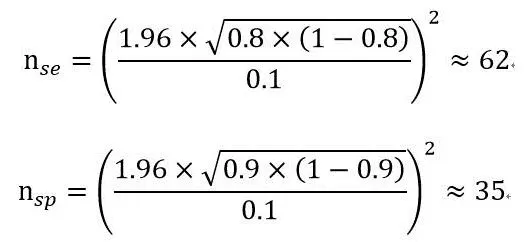
We then calculate the two sample sizes based on sensitivity and specificity, respectively, the larger of which is the sample size required. In this case, the sample size calculated by the formula is 62 cases.
We can also calculate the sample size directly using the PASS software:


The results of this example show that N=70 by sensitivity; N=44 by specificity, the larger value is to be selected; that is, 70 cases of research subjects are needed.
The results of PASS software calculations are slightly deviated from the formula calculations, and the results with a larger sample size can be selected, i.e., the required sample size is 70 cases for the control group and 70 cases for the case group.
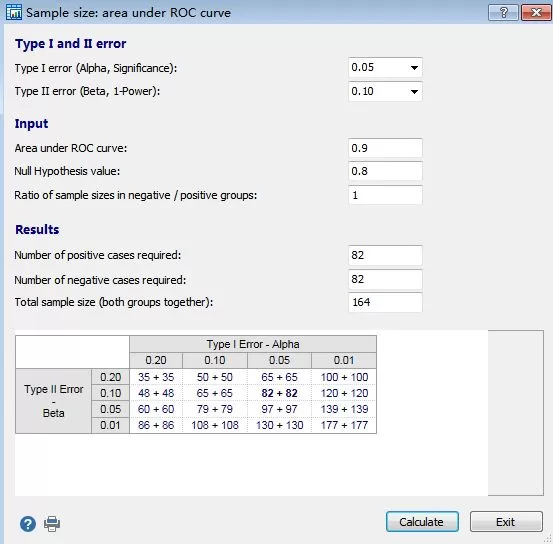
Calculation according to AUC value
 Software: MedCalc
Software: MedCalc
For example, if we set the parameters to:
Expected AUC=0.9
Test level α: 0.05
Test efficacy 1-β: 0.9
Null hypothesis: AUC=0.8
In machine learning, the ratio of the training set, validation set, and test set is generally taken as 6:2:2. If there is no validation set, the ratio of the training set to test set is cut at 7:3. When mapped to the metabolic marker screening process, this corresponds to a 7:3 ratio of the modeling cohort to the validation cohort.
2. Developing sample inclusion/exclusion criteria
Metabolomics results are susceptible to external influences (such as the environment). Therefore, strict inclusion criteria for samples must be applied to screen for true-positive metabolic biomarkers.
Sample inclusion criteria
(1) Randomly select samples that meet the grouping requirements as per the experimental design.
(2) Clinical indicators such as age, gender, and others not relevant to the study should not differ between groups as much as possible.
(3) Sample selection should be as random as possible in terms of time and geography.
(4) Samples included should comply with the standard sampling procedure and with complete corresponding clinical information.
Common clinical information to be collected is listed below:
|
Routine clinical information |
|||||||
|
Sample number |
Name |
Gender |
Age |
Ethnicity |
Body Height |
Body Weight |
BMI |
|
Blood pressure |
Alcohol consumption history |
Smoking history |
Disease history |
Residency |
Blood counts |
Biochemical parameters |
etc. |
|
Specific clinical information |
|||||||
|
Disease type |
Disease subtype |
Disease staging |
Disease markers |
"Gold standard" |
etc. |
|
|
Sample exclusion criteria
(1) Samples with severe acute infections;
(2) Samples with severe anemia;
(3) Samples with hepatic or renal insufficiency;
(4) Samples with a tumor or multiple tumors;
(5) Samples in pregnancy;
(6) Samples with autoimmune deficiency diseases;
(7) Samples with hyperlipidemia or hyperjaundice.
The above exclusions only apply to studies that are not specific to that disease/condition (i.e., the 5th exclusion does not apply in the case of studies related to gestational diabetes mellitus).
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


