What is Protein Sequencing: Significance, Methods, and Applications
Protein sequencing is a cornerstone of modern biological research, providing crucial insights into the structure and function of proteins. By determining the precise order of amino acids in a protein, researchers can unlock the secrets of cellular processes, disease mechanisms, and potential therapeutic targets. Whether you are involved in drug development, structural biology, or personalized medicine, understanding protein sequencing is essential. This article delves into the significance of protein sequencing, the methods used, and its diverse applications in various scientific fields.
- What is Protein Sequencing?
- Methods and Techniques for Protein Sequencing
- Applications of Protein Sequencing
- Challenges and Solutions of Protein Sequencing
- Technological Advancements of Protein Sequencing
- How does mass spectrometry contribute to protein sequencing?
- Bioinformatics and Data Analysis
1. What is Protein Sequencing?
Protein sequencing is a critical process in the field of biochemistry and molecular biology, aimed at determining the amino acid sequence of proteins. This technique is essential for understanding the structure, function, and interactions of proteins, which are vital macromolecules in all living organisms. By identifying the precise order of amino acids, scientists can gain insights into the protein's role within the cell, how it interacts with other molecules, and how alterations in its sequence can lead to various diseases.
The Significance of Protein Sequencing
The significance of protein sequencing extends far beyond basic scientific research. For instance, in the pharmaceutical industry, understanding the exact sequence of proteins can lead to the development of new drugs and therapeutic strategies. By targeting specific proteins associated with diseases, researchers can design molecules that can either inhibit or enhance the protein's function, offering new treatments for conditions such as cancer, Alzheimer's, and diabetes.
Moreover, protein sequencing plays a crucial role in the field of biotechnology. It allows for the engineering of proteins with desirable traits, which can be used in various industrial applications, from producing biofuels to developing new materials. In agriculture, protein sequencing helps in improving crop resistance to pests and diseases, thereby enhancing food security.
2. Methods and Techniques for Protein Sequencing
Protein sequencing has evolved significantly over the years, with various methods and techniques being developed to determine the amino acid sequences of proteins. Understanding these techniques is crucial for researchers and industry professionals who rely on accurate protein sequencing for their work. Here, we explore three primary methods: Edman Degradation, Mass Spectrometry, and Next-Generation Sequencing.
Edman Degradation
One of the earliest methods for protein sequencing is Edman degradation. This method involves the stepwise removal and identification of the N-terminal amino acid of a protein. The process uses phenyl isothiocyanate to label the N-terminal residue, which is then cleaved and identified through chromatography. While Edman degradation was revolutionary at its inception, it has several limitations. For instance, it requires relatively large quantities of pure protein and can struggle with proteins that have repetitive sequences or post-translational modifications.
Mass Spectrometry
Mass spectrometry has emerged as a powerful tool for protein sequencing. This technique involves ionizing protein fragments and measuring their mass-to-charge ratios. Modern mass spectrometry techniques, such as tandem mass spectrometry (MS/MS), enable the identification of peptides and their sequences with high accuracy. Mass spectrometry offers high sensitivity and can handle complex mixtures of proteins, making it suitable for analyzing samples with diverse and low-abundance proteins. However, it may face challenges with membrane proteins and requires sophisticated equipment and expertise.
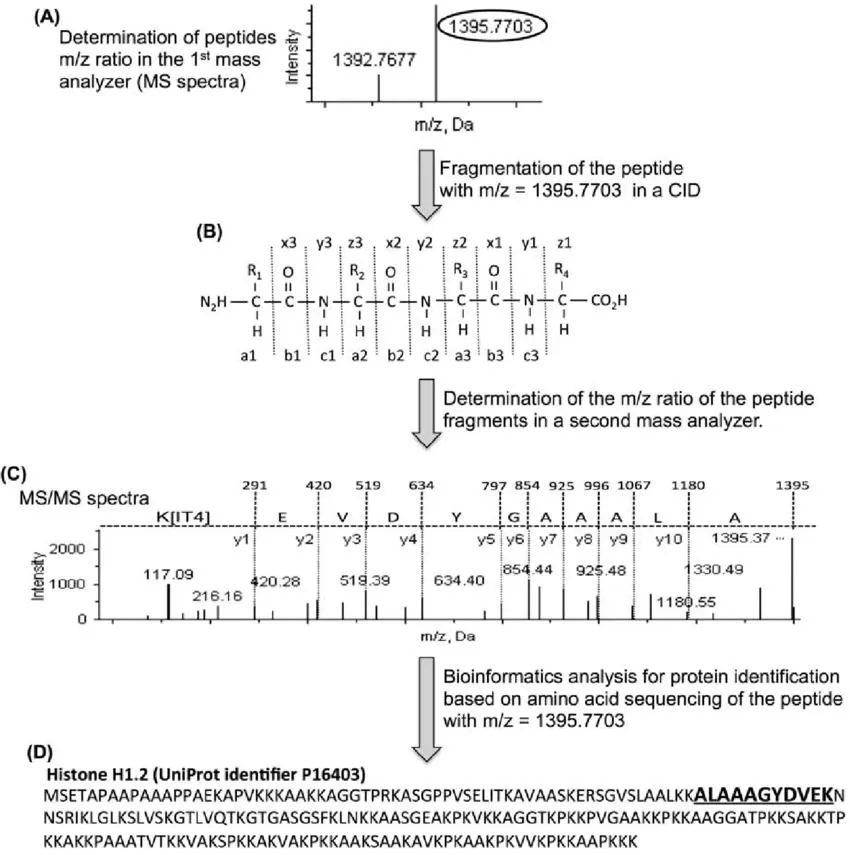
Next-Generation Sequencing
Recent advancements in next-generation sequencing (NGS) technologies have expanded their application beyond genomics to proteomics. NGS-based methods, such as RNA-seq and ribosome profiling, can indirectly infer protein sequences by analyzing the corresponding mRNA sequences. These techniques offer high-throughput capabilities and can provide comprehensive insights into the proteome. However, they rely on mRNA data and may not always provide direct protein sequence information, which can be a limitation when studying proteins with complex post-translational modifications or those that undergo alternative splicing.
3. Applications of Protein Sequencing
Protein sequencing is a powerful tool that has revolutionized various fields of science and industry. From drug development to personalized medicine, the applications of protein sequencing are vast and impactful. In this section, we will explore how protein sequencing is applied in drug development, structural biology, proteomics research, biotechnology, and biopharmaceuticals, as well as personalized medicine.
Drug Development
Protein sequencing is instrumental in drug discovery and development. By elucidating the primary structure of target proteins, researchers can design molecules, such as small-molecule drugs or biologics, that specifically interact with these proteins. This targeted approach forms the basis for developing novel therapeutics. For example:
Targeted Cancer Therapies: Sequencing proteins involved in cancer pathways, like tyrosine kinases, has led to the development of drugs like imatinib (Gleevec) for chronic myeloid leukemia.
Monoclonal Antibodies: Monoclonal antibodies used in immunotherapy are often designed based on the recognition of specific protein epitopes. These antibodies can precisely target cancer cells, leading to more effective treatments with fewer side effects.
Structural Biology
Understanding the primary structure of proteins is a prerequisite for elucidating their three-dimensional structures. Techniques like X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, which rely on the known amino acid sequence, allow scientists to visualize the protein's shape. This knowledge is invaluable for designing drugs that target specific protein conformations. By knowing the precise arrangement of amino acids, researchers can predict how a protein will fold, what its active sites might be, and how it might interact with other molecules.
Proteomics Research
In the realm of proteomics, protein sequencing serves as the foundational step. By sequencing proteins in a given biological sample, researchers gain insights into cellular processes, identify biomarkers, and uncover the mechanisms underlying various diseases. This holistic approach has led to numerous breakthroughs:
Cancer Proteomics: Profiling the proteome of cancer cells has revealed unique protein signatures and potential drug targets, enabling the development of more precise and effective cancer therapies.
Neuroproteomics: Sequencing neuronal proteins aids in understanding neurological disorders and neurodegenerative diseases, paving the way for new treatments and diagnostic tools for conditions like Alzheimer's and Parkinson's diseases.
Functional Proteomics: Studying protein interactions, post-translational modifications, and expression patterns enhances our comprehension of cellular function and helps identify potential therapeutic targets for various diseases.
Biotechnology and Biopharmaceuticals
In biotechnology, protein sequencing is indispensable for developing and manufacturing biopharmaceuticals, recombinant proteins, and enzymes. By sequencing and engineering proteins, researchers can create molecules tailored for specific functions, including:
Enzyme Engineering: Altering the sequences of enzymes can enhance their catalytic efficiency and substrate specificity for various industrial applications, such as biofuel production, food processing, and waste management.
Biologics Production: Protein sequencing informs the design and optimization of therapeutic monoclonal antibodies and other biologics, ensuring their efficacy and safety in treating diseases like cancer, autoimmune disorders, and infectious diseases.
Personalized Medicine
The era of personalized medicine relies on genetic and proteomic data to tailor medical treatments to individual patients. Protein sequencing plays a crucial role in this paradigm by identifying patient-specific genetic mutations and variations, thereby enabling the development of personalized treatment strategies. By understanding the unique protein sequences present in an individual's cells, healthcare providers can design customized therapies that are more effective and have fewer side effects.
4. Challenges and Solutions of Protein Sequencing
While protein sequencing has revolutionized many fields, it comes with its own set of challenges. These challenges span various stages of the sequencing process, from sample preparation to data analysis and cost. However, with continuous advancements in technology and methodology, solutions are being developed to overcome these hurdles. Let's explore some of the main challenges in protein sequencing and the solutions that are being implemented.
Sample Preparation
Sample preparation is a critical but often challenging step in protein sequencing. Proteins must be extracted, purified, and sometimes modified before sequencing can occur. This process can be complex and time-consuming, with each type of sample requiring specific handling protocols to ensure the integrity and purity of the proteins. Solutions to these challenges include:
Improved Sample Handling Techniques: Developing more robust and versatile sample handling techniques helps in maintaining the quality of proteins during extraction and purification. Techniques such as microfluidic devices and automated sample preparation systems are being used to enhance efficiency and reduce the risk of contamination.
Sample Preparation Kits: The development of specialized kits for sample preparation has streamlined the process, making it more accessible and standardized. These kits are designed to handle various types of samples, ensuring that proteins are adequately prepared for sequencing.
Data Analysis
Analyzing the vast amount of data generated during protein sequencing can be daunting. The complexity of the data requires sophisticated tools to accurately interpret and identify the sequences. Advanced bioinformatics tools and software packages have been developed to automate data analysis, simplifying the identification of peptides and proteins from mass spectrometry data. Solutions include:
Advanced Bioinformatics Tools: These tools can process large datasets, identify patterns, and make sense of complex sequencing data. Software such as MaxQuant and Proteome Discoverer are widely used in the field to facilitate protein data analysis.
Automated Data Analysis Pipelines: By automating various steps of data analysis, these pipelines reduce human error and increase throughput. They allow researchers to quickly and accurately analyze sequencing data, leading to faster and more reliable results.
Cost
Cost can be a limiting factor, especially for large-scale proteomic studies. Protein sequencing requires expensive reagents, equipment, and software, making it a significant investment for many laboratories. To address this challenge, researchers are continually working on cost-effective sequencing techniques and optimizing workflows to maximize data yield while minimizing expenses. Solutions include:
Cost-Effective Sequencing Techniques: Innovations in sequencing technology are driving down costs. Methods such as label-free quantification and high-throughput mass spectrometry are becoming more affordable, making protein sequencing more accessible to a broader range of researchers.
Optimized Workflows: By streamlining the sequencing process, researchers can reduce the time and resources required. This includes optimizing sample preparation, data acquisition, and analysis steps to ensure maximum efficiency and cost-effectiveness.
Collaborative Research: Sharing resources and collaborating with other research institutions can help mitigate costs. By pooling resources, laboratories can afford the necessary equipment and reagents for protein sequencing projects.
5. Technological Advancements of Protein Sequencing
The field of protein sequencing has witnessed remarkable technological advancements over the past few decades. These innovations have significantly enhanced the speed, accuracy, and efficiency of protein sequencing, paving the way for groundbreaking discoveries in proteomics, drug development, and personalized medicine. Let's delve into some of the key technological advancements in protein sequencing.
High-Throughput Sequencing
Recent technological advancements have led to the development of high-throughput protein sequencing techniques. These methods allow the simultaneous analysis of thousands of proteins, significantly accelerating the pace of proteomics research. High-throughput sequencing technologies have transformed our understanding of complex biological systems by enabling large-scale protein profiling.
Automation and Miniaturization: Advances in automation and miniaturization have made it possible to conduct high-throughput protein sequencing efficiently. Automated systems reduce human error and increase throughput, while miniaturization allows for the analysis of smaller sample volumes, making the process more cost-effective.
Multiplexing: Multiplexing techniques enable the simultaneous sequencing of multiple proteins in a single experiment. This approach maximizes the amount of data obtained from a single run, enhancing the overall efficiency of protein sequencing projects.
Mass Spectrometry Improvements
Mass spectrometry has been a cornerstone of protein sequencing for many years, and recent improvements have further solidified its importance. Modern mass spectrometry instruments are more sensitive and precise, allowing for the detection of low-abundance proteins and post-translational modifications with greater accuracy.
Enhanced Sensitivity: Improvements in mass spectrometry sensitivity have made it possible to detect proteins present in very low concentrations. This capability is crucial for studying proteins involved in signaling pathways and other cellular processes where protein abundance is typically low.
Advanced Data Analysis Software: Innovations in data analysis software have significantly improved the accuracy of protein identification. Tools such as MaxQuant and Proteome Discoverer use sophisticated algorithms to analyze mass spectrometry data, providing more reliable and comprehensive protein identification.
Tandem Mass Spectrometry (MS/MS): Tandem mass spectrometry, or MS/MS, involves multiple rounds of mass spectrometry analysis. This technique provides detailed information about protein sequences and post-translational modifications, enhancing the overall quality of protein sequencing data.
Hybrid Approaches
Hybrid approaches that combine different sequencing methods have emerged as powerful tools for protein sequencing. By integrating techniques such as Edman degradation and mass spectrometry, researchers can achieve enhanced sequencing capabilities and accuracy.
Combination of Methods: Combining different sequencing methods allows researchers to leverage the strengths of each technique. For instance, Edman degradation can provide detailed N-terminal sequencing information, while mass spectrometry can identify internal and C-terminal sequences.
Cross-Validation: Hybrid approaches enable cross-validation of sequencing data, increasing confidence in the results. By confirming findings using multiple methods, researchers can ensure the accuracy and reliability of their protein sequencing data.
Enhanced Coverage: Hybrid methods offer improved coverage of protein sequences, including challenging regions such as repetitive sequences and post-translational modifications. This comprehensive approach provides a more complete understanding of protein structure and function.
6. How Does Mass Spectrometry Contribute to Protein Sequencing?
Mass spectrometry (MS) has emerged as a cornerstone technology in the field of protein sequencing, providing unparalleled sensitivity, accuracy, and throughput. This powerful analytical technique has revolutionized our ability to study proteins, identify their sequences, and understand their functions. Here’s a detailed look at how mass spectrometry contributes to protein sequencing.
Principles of Mass Spectrometry
Mass spectrometry works by ionizing protein molecules, fragmenting them into smaller peptides, and measuring their mass-to-charge (m/z) ratios. The resulting data provides a "mass spectrum" that can be used to infer the sequence of the protein. The process generally involves three main steps:
1. Ionization: Proteins are ionized using techniques such as Electrospray Ionization (ESI) or Matrix-Assisted Laser Desorption/Ionization (MALDI). These methods convert protein molecules into charged ions.
2. Mass Analysis: The ions are then passed through a mass analyzer, which separates them based on their m/z ratios. Common mass analyzers include Time-of-Flight (TOF), Quadrupole, and Ion Trap analyzers.
3. Detection: The separated ions are detected, and their abundances are recorded, generating a mass spectrum. This spectrum serves as a fingerprint of the protein, which can be analyzed to determine its sequence.
Tandem Mass Spectrometry (MS/MS)
Tandem mass spectrometry (MS/MS) enhances the capabilities of traditional MS by adding an additional stage of fragmentation and analysis. In MS/MS, protein ions are fragmented into smaller peptide ions, which are then subjected to a second round of mass spectrometry. This process provides detailed sequence information and improves the accuracy of protein identification.
Fragmentation Methods: Common fragmentation methods used in MS/MS include Collision-Induced Dissociation (CID), Higher-energy Collisional Dissociation (HCD), and Electron Transfer Dissociation (ETD). These techniques generate different types of peptide fragments, offering complementary sequence information.
Peptide Sequencing: MS/MS data is used to generate peptide mass fingerprints, which are compared against protein databases to identify the protein and determine its sequence. This approach is highly effective for identifying proteins in complex mixtures.
Post-Translational Modifications (PTMs)
Mass spectrometry is particularly valuable for studying post-translational modifications (PTMs), which are critical for regulating protein function. PTMs such as phosphorylation, glycosylation, and ubiquitination can be identified and characterized using MS techniques.
Sensitivity and Specificity: Advanced MS techniques can detect and quantify PTMs with high sensitivity and specificity. This capability is crucial for understanding how PTMs affect protein activity, interactions, and cellular localization.
Site Localization: MS can also pinpoint the exact amino acid residues that are modified, providing insights into the functional consequences of PTMs. This information is essential for understanding protein regulation and developing targeted therapeutics.
Quantitative Proteomics
Mass spectrometry enables quantitative proteomics, allowing researchers to measure protein abundance and compare expression levels across different samples. Techniques such as Isobaric Tag for Relative and Absolute Quantitation (iTRAQ) and Tandem Mass Tags (TMT) facilitate accurate quantification of proteins in complex biological samples.
Label-Free Quantification: In addition to labeled quantification methods, label-free quantification techniques are also used. These methods rely on the intensity of peptide signals to estimate protein abundance, offering a cost-effective alternative for large-scale studies.
Comparative Analysis: Quantitative proteomics enables the comparison of protein expression profiles between different conditions, such as healthy and diseased states. This comparative approach helps identify biomarkers and therapeutic targets.
Advantages and Limitations
Mass spectrometry offers several advantages for protein sequencing, including high sensitivity, specificity, and the ability to analyze complex mixtures. However, it also has some limitations that must be considered.
Advantages:
- High Sensitivity: Capable of detecting low-abundance proteins and rare PTMs.
- Versatility: Applicable to a wide range of proteins and sample types.
- High Throughput: Enables large-scale proteomic studies and comprehensive protein profiling.
Limitations:
- Sample Preparation: Requires careful and sometimes complex sample preparation.
- Data Complexity: Generates large datasets that require advanced bioinformatics tools for analysis.
- Cost: High-quality MS instruments and reagents can be expensive, limiting accessibility for some labs.
7. Bioinformatics and Data Analysis
In the age of vast data, bioinformatics assumes a crucial role in the domain of protein sequencing. Advanced bioinformatics tools and software applications are indispensable for analyzing the massive amounts of data generated from protein sequencing experiments. These tools help in identifying proteins, quantifying their abundance, and interpreting their biological significance.
Key Software Applications
Software applications such as MaxQuant, Proteome Discoverer, and Mascot are pivotal in the field of protein sequencing. These tools streamline data analysis by matching experimental data against comprehensive protein sequence databases. This process enables the accurate identification of proteins and their post-translational modifications (PTMs), which are crucial for understanding protein function and regulation.
MaxQuant
MaxQuant is a widely used software for mass spectrometry-based proteomics. It offers robust algorithms for identifying and quantifying proteins, analyzing PTMs, and providing statistical validation of the results. MaxQuant's integration with the Andromeda search engine enhances its ability to handle complex datasets and large-scale proteomic studies.
Proteome Discoverer
Proteome Discoverer is another powerful tool designed to analyze mass spectrometry data. It supports a wide range of workflows for protein identification, PTM analysis, and quantification. Its modular architecture allows researchers to customize their analyses according to specific experimental needs, making it highly versatile in proteomics research.
Mascot
Mascot is a popular search engine for identifying proteins by matching mass spectrometry data to protein sequence databases. It provides high sensitivity and specificity in protein identification and is widely used for both basic research and clinical proteomics. Mascot's ability to handle large datasets efficiently makes it a valuable asset in the bioinformatics toolkit.
Machine Learning in Bioinformatics
The integration of machine learning algorithms in bioinformatics is revolutionizing the field of protein sequencing. These algorithms enhance the precision of protein identification and PTM analysis by learning from vast amounts of experimental data. Machine learning models can predict protein structures, interactions, and functional annotations, providing deeper insights into proteomics.
Predictive Modeling
Predictive modeling uses machine learning to anticipate protein structures and interactions based on sequence data. These models are trained on known protein structures and sequences, enabling them to make accurate predictions for newly sequenced proteins. This approach accelerates the discovery of protein functions and potential therapeutic targets.
PTM Analysis
Machine learning algorithms are also employed to detect and analyze PTMs, which are crucial for understanding protein regulation and function. By analyzing patterns in mass spectrometry data, these algorithms can identify novel PTMs and provide insights into their biological roles, thereby advancing our knowledge of cellular processes.
Partner with MetwareBio for Advanced Proteomics Solutions
Protein sequencing has revolutionized the way we understand biological processes and develop innovative solutions in healthcare and biotechnology. As we continue to advance in this field, the expertise of specialized service providers becomes invaluable. MetwareBio, a leading proteomics service provider, offers comprehensive solutions to meet your research needs. Explore their DIA Quantitative Proteomics services and gain unparalleled insights into protein dynamics. Visit MetwareBio today to learn more about how their cutting-edge technologies can drive your research forward.
References
- Alsagaby, Suliman A. "Understanding the fundamentals of proteomics." Curr Top Pept Protein Res 20.3 (2019): 25-33.
- Graves, Paul R., and Timothy AJ Haystead. "Molecular biologist's guide to proteomics." Microbiology and molecular biology reviews 66.1 (2002): 39-63.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


