Unlocking Proteogenomics: Bridging Genomics and Proteomics
Introduction to Proteogenomics
Proteogenomics is the intersection of proteomics and genomics, integrating genomic, transcriptomic, proteomic, and post-translational modification data to systematically detect molecular changes in biological samples, providing a comprehensive perspective through multi-omics analysis.
Significant Research: Pear Proteogenomic Atlas
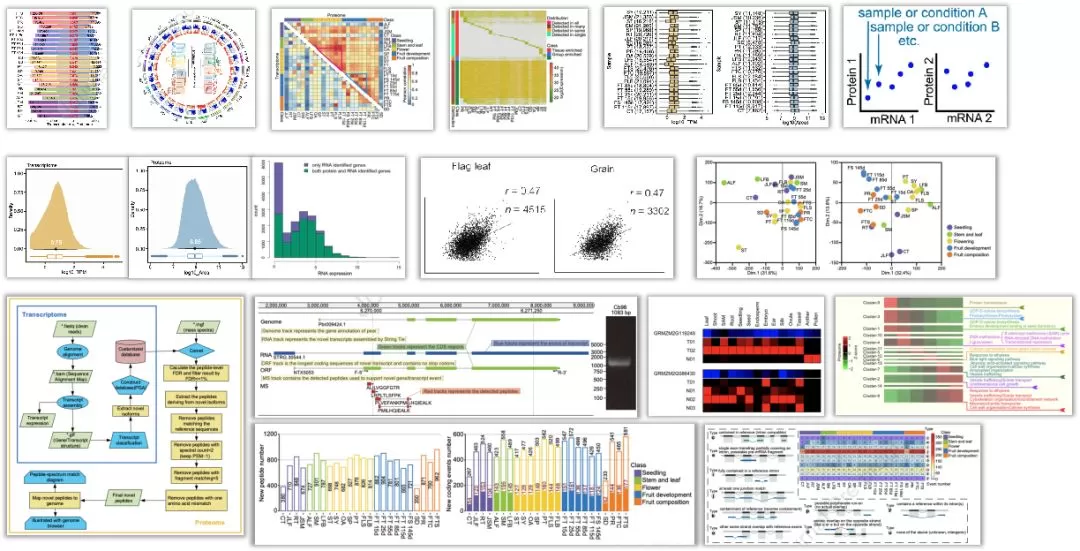
In 2023, the journal Molecular Plant published a paper titled "A large-scale proteogenomic atlas of pear", which conducted protein-genome research on 24 pear tissue organs, thereby refining the pear genome annotation and exploring key genes related to important agronomic traits.
The Role of Shotgun Proteomics in Proteogenomics
Proteomics typically employs a "shotgun proteomics" approach, wherein proteins extracted from samples are enzymatically digested into peptides. These peptides are then analyzed using liquid chromatography-mass spectrometry (LC-MS) to obtain peptide mass spectrometry information, facilitating qualitative analysis of peptides and proteins by matching the peptide tandem mass spectra with theoretical spectra of candidate peptides in protein sequence databases.
Challenges in Protein Sequence Databases
The depth of protein detection in proteomics studies heavily relies on the completeness of protein sequence databases. Many peptides do not exist in specific reference protein sequence databases or any reference databases, leading to the omission of numerous protein information. The majority of information in protein sequence databases originates from genome-predicted amino acid sequences (based on the open reading frame principle), thus the degree of genome annotation directly affects the completeness of protein sequence databases.
Integration of Genomic and Proteomic Data
In protein-genome research, custom protein sequence databases generated using genomic and transcriptomic information are used to aid in identifying new peptides (which do not exist in reference protein sequence databases) from mass spectrometry-based proteomics data. Conversely, proteomics data can provide evidence of gene expression at the protein level and help refine genome annotation by identifying new transcripts.
_1721379120_WNo_865d621.webp)
Methodology: Creating Custom Protein Sequence Databases
Protein-genome research essentially involves the integration of multi-omics data, wherein a comprehensive protein sequence database is constructed using genomic and transcriptomic data, followed by validation of the reliability of new transcripts through proteomic data and annotation of the genome in reverse. To obtain as much proteomic and transcriptomic data as possible, it is recommended to collect samples from multiple tissues and time periods for proteomic and transcriptomic detection. Basic analysis of proteomic and transcriptomic data helps understand the characteristics of whole tissue/developmental stage sample data, followed by assembly and prediction of new coding events in transcriptomic data. A more comprehensive protein sequence database is constructed based on existing data, and this new database is used to identify peptide segments in proteomic data. New peptide segments and new coding events are then used to refine genome annotation. Gene function analysis is performed based on refined proteomic and transcriptomic data to explore functional genes.
Modules of Protein-Genome Analysis
Protein-genome analysis consists of three main modules: omics data comparative analysis, annotation of new peptide segments/transcripts, and gene function analysis. Omics data comparative analysis enables quality control of overall data and explores potential post-transcriptional regulation mechanisms by mining transcription and protein level expression differential genes. Annotation of new peptide segments/transcripts discovers new coding events, improves genome annotation, and facilitates the discovery of new mechanisms. Gene function analysis associates genes with phenotypes, effectively screening candidate genes and performing functional annotation.
Conclusion: The Future of Proteogenomics
Proteogenomics offers a powerful approach to uncovering new insights into molecular biology by bridging the gap between genomics and proteomics. As methodologies advance and databases become more comprehensive, proteogenomics will continue to play a crucial role in scientific research and discovery.
Reference:
Armengaud J, Trapp J, Pible O, Geffard O, Chaumot A, Hartmann EM. Non-model organisms, a species endangered by proteogenomics. J Proteomics. 2014;105:5-18. doi:10.1016/j.jprot.2014.01.007
Read more:
· A Guide to Protein Database Selection
· MetwareBio Launches Proteomics Services
· What is Isoelectric Points of Amino Acids: Essential Calculations and Practical Applications
· Comparison and Application of Proteomic Technologies
· Demystifying Proteomics Research Strategies and Content in a Single Read
· Optimal Protein Database Selection: Insights from Experimental Data
· Protein sample preparation tips: Serum or Plasma?
· An Overview of Mainstream Proteomics Techniques
· Exploring Disease Mechanisms: Key Factors in Proteomic Analysis
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


