Ubiquitination and Phosphorylation: The Dynamic Duo of Cellular Regulation
Chemical Process of Protein Phosphorylation and Ubiquitination
Protein post-translational modifications (PTMs) refer to chemical alterations that occur at specific sites on a protein molecule. Common types of PTMs include phosphorylation, glycosylation, acetylation, and ubiquitination. These modifications regulate various biological processes, such as signal transduction and cell differentiation, by altering the activity, localization, folding, and function of proteins. Among the many types of PTMs, ubiquitination, phosphorylation, and acetylation are the most extensively studied.
Protein phosphorylation is the process in which a phosphate group from ATP or GTP is transferred to the amino acid residues of a target protein (such as serine, threonine, or tyrosine) by protein kinases. Reversible protein phosphorylation regulates nearly every aspect of cellular processes, including replication, transcription, apoptosis, immune responses, environmental adaptation, and cellular metabolism. Abnormal phosphorylation has been linked to various diseases, including cancer, cardiovascular disorders, and metabolic syndromes. As a result, drugs targeting protein kinases have become attractive therapeutic strategies for several diseases.
Protein ubiquitination involves the attachment of ubiquitin (Ub), a small protein consisting of 76 amino acids, to target proteins through a series of enzymatic reactions, forming a conserved post-translational modification mechanism. This process involves three key enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). Ubiquitin can be attached to substrates in various ways, including the addition of a single ubiquitin molecule to a single lysine residue (monoubiquitination), multiple ubiquitin molecules attached to multiple lysine residues (multi-monoubiquitination), or the formation of ubiquitin chains (polyubiquitination). These ubiquitin chains are linked to the substrate protein's N-terminal methionine (M1) or one of seven internal lysine residues (K6, K11, K27, K29, K33, K48, K63), resulting in different structural configurations. Both monoubiquitination and polyubiquitination play critical roles in regulating protein function under both physiological and pathological conditions.
Molecular Regulatory Mechanisms of Ubiquitination and Phosphorylation
Ubiquitination and phosphorylation play crucial regulatory roles in various cellular signaling pathways. Their specific mechanisms are outlined as follows:
I. Phosphorylation:
a) Phosphorylation is a reversible and dynamic process regulated by the competing activities of protein kinases and phosphatases, impacting multiple intracellular signaling pathways. It often serves as a key mechanism for activating essential regulatory proteins and controlling signal transduction.
b) In the mTOR complex 1 (TORC1) signaling pathway, both phosphorylation and ubiquitination modifications play significant roles in transmitting external signals to TORC1 and regulating its downstream pathways.
c) Phosphorylation also participates in the negative regulation of the androgen receptor signaling pathway in prostate cancer cells, downregulating the transcriptional activity of the androgen receptor through PAK6-mediated phosphorylation-dependent ubiquitination.
II. Ubiquitination:
a) Ubiquitination involves the covalent attachment of the small ubiquitin molecule to target proteins, regulating protein degradation and signal transduction. The interactions within the ubiquitin-proteasome system (UPS) create a complex ubiquitination signaling network that is integral to regulating cellular activities, including the cell cycle, immune response, and signaling pathways.
b) Similarly, in the mTOR complex 1 (TORC1) signaling pathway, ubiquitination modifications significantly influence the pathway's transmission and regulation.
c) Ubiquitination also controls the activity of NF-κB, modulating the signaling pathways involved in the responses of pro-inflammatory cytokines, such as tumor necrosis factor-alpha (TNF-α) and interleukin-1, through ubiquitin-dependent proteolysis.
The specific mechanisms of ubiquitination and phosphorylation in various cellular signaling pathways include: phosphorylation activating key regulatory proteins and controlling signal transduction through dynamic regulation by protein kinases and phosphatases, while ubiquitination regulates protein degradation and signal transduction through the covalent attachment of ubiquitin molecules to target proteins, thus participating in the regulation of diverse cellular functions.
Synergistic Regulation of Ubiquitination and Phosphorylation
Ubiquitination and phosphorylation are critical mechanisms that regulate protein degradation, precisely controlling intracellular signaling pathways and protein stability. In eukaryotic cells, ubiquitination primarily occurs through the ubiquitin-proteasome system (UPS), which relies on a cascade of reactions involving ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) to attach ubiquitin molecules to target proteins. Once a protein is ubiquitinated, it is recognized by the proteasome and subsequently degraded.
In some cases, phosphorylation can enhance or inhibit the ubiquitination process. Specifically, phosphorylation may affect the selective recognition of substrates by E3 ubiquitin ligases or directly participate in cross-linking reactions. Furthermore, phosphorylation can create specific phosphodegrons, making substrate proteins more susceptible to ubiquitination and degradation. Research has revealed complex interactions between phosphorylation and ubiquitination. For instance, within the mammalian TORC1 signaling pathway, phosphorylation and ubiquitination work closely together, with some substrate proteins requiring phosphorylation prior to ubiquitination. Similarly, in rice immune responses, the levels and activity of the WRKY45 transcription factor are co-regulated by PUB44-mediated ubiquitination and MPK6-mediated phosphorylation.
Importantly, the interplay between different types of post-translational modifications enhances the specificity and combinatorial logic of signaling. For example, O-GlcNAc modification competes with phosphorylation during the post-translational modification process, and both often coexist on the same protein molecule. This competitive relationship among modifications further complicates the mechanisms of protein degradation.
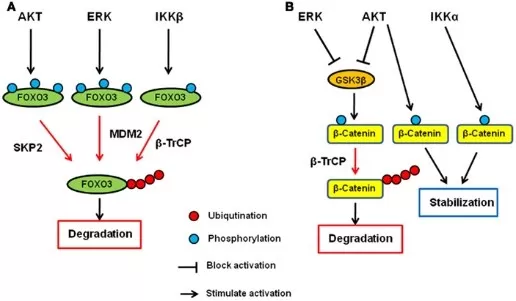
The synergy between ubiquitination and phosphorylation not only ensures the precision and specificity of protein degradation but also equips cells with the ability to respond to environmental changes and regulate gene expression. These mechanisms play a vital role in maintaining cellular homeostasis and modulating the progression of diseases.
Reference
1. Xiumei Wu,Mengyun Xu,Mengya Geng,et al. Targeting protein modifications in metabolic diseases: molecular mechanisms and targeted therapies. Signal Transduction and Targeted Therapy,8(1),2023.
2. Xinyan Chen,Anat Raiff,Shanshan Li,et al. Mechanism of Ψ-Pro/C-degron recognition by the CRL2(FEM1B) ubiquitin ligase.Nature Commmuication,15(2),2024.
3. Ping Liu,Xiaoji Cong,Shengjie Liao,et al. Global identification of phospho-dependent SCF substrates reveals a FBXO22 phosphodegron and an ERK-FBXO22-BAG3 axis in tumorigenesis.Cell Death Differentiation,29(1),2022.
4. Kota Ichimaru,Koji Yamaguchi,Kenichi Harada,et al. Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity. Nature Commmuication,13(1),2022.
5. Danielle Swaney,Pedro Beltrao,Lea Starita, et al. Global analysis of phosphorylation and ubiquitylation crosstalk in protein degradation.Nature Methods,10(7),2013.
6. Yamaguchi H, Hsu JL and Hung M-C (2012) Regulation of ubiquitination-mediated protein degradation by survival kinases in cancer. Front. Oncol. 2:15. doi: 10.3389/fonc.2012.00015


