Decoding Protein Interaction Networks: Advanced Strategies in Tandem Affinity Purification (TAP)
Tandem Affinity Purification (TAP) is a powerful method for isolating native protein complexes through two sequential affinity purification steps. By drastically reducing nonspecific binding, TAP enables systematic exploration of protein interaction networks and has become a gold-standard technique for studying macromolecular assemblies. This blog outlines the principles, workflow, and comparative advantages of TAP, while highlighting its critical role in modern proteomics.
1. Principles of TAP
TAP relies on the sequential use of two distinct affinity tags to purify target proteins along with their interacting partners under native conditions. This approach ensures high specificity and minimizes contamination from nonspecific interactions.
Key Steps in TAP Workflow:
1) Tag Fusion: The gene encoding the target protein is fused with two orthogonal affinity tags (e.g., Protein A and Calmodulin-Binding Peptide).
2) Expression: The tagged protein is expressed in a suitable host system (e.g., mammalian cells, yeast), allowing it to form native complexes.
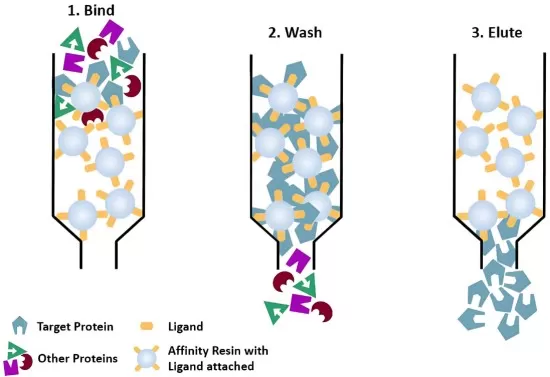
3) First Purification: The cell lysate is subjected to the first affinity purification step using a resin specific to Tag 1 (e.g., IgG Sepharose for Protein A). Harsh wash conditions (e.g., high salt, detergents) remove loosely bound contaminants.
4) Tag Cleavage: A protease (e.g., TEV) cleaves the first tag, releasing the protein complex.
5) Second Purification: The eluate undergoes a second affinity purification using Tag 2 (e.g., Calmodulin resin for CBP). Mild elution conditions (e.g., EGTA-mediated calcium chelation) preserve complex integrity.
6) Analysis: Purified complexes are analyzed via mass spectrometry (MS) or Western blotting to identify interaction partners.
Affinity Purification
2. Core Design and Optimization of TAP
2.1 Dual-Tag System
The choice of tags is critical for TAP success:
Tag 1: High-affinity and robust under stringent conditions.
- Protein A: Binds IgG resin but may sterically hinder protein function due to its large size (~14 kDa).
- Strep-tag II: Small (8 aa), binds Strep-Tactin resin, and allows gentle elution via biotin competition.
Tag 2: Orthogonal to Tag 1 with mild elution.
- CBP: Binds calmodulin resin in a calcium-dependent manner; eluted via EGTA.
- FLAG-tag: Recognized by Anti-FLAG M2 resin; eluted using FLAG peptide.
2.2 Expression Systems
Mammalian Cells (HEK293T, HeLa):
- Ideal for human proteins requiring post-translational modifications.
- Use weak promoters (e.g., EF1α) or inducible systems (e.g., TetOn) to avoid overexpression artifacts.
Yeast (S. cerevisiae):
- Genome integration via homologous recombination (e.g., pBS1479 vector).
- Inducible expression using galactose promoters (e.g., GAL1/10).
Bacteria/Insect Cells: Less common due to challenges in preserving native complex structures.
3. Experimental Protocol for TAP
3.1 Plasmid Construction and Validation
- Primer Design: Insert TAP tags at the Nor C-terminus, ensuring proper reading frames.
- Cloning Verification: Confirm tag insertion via sequencing and validate fusion protein expression using Western blot (e.g., anti-Protein A antibodies).
- Negative Controls: Include empty vectors or tag-only constructs to rule out nonspecific interactions.
3.2 Cell Transfection and Induction
- Mammalian Cells: Transfect using PEI or liposomes (e.g., Lipofectamine 3000); harvest cells after 72 hours.
- Stable Line Selection: Apply puromycin (2–5 μg/mL) for 1–2 weeks.
- Inducible Systems: Induce expression with doxycycline (1 μg/mL) for 24 hours.
3.3 Cell Lysis and Preprocessing
- Lyse cells on ice for 30 minutes, followed by sonication (3 × 10-second pulses at 30% amplitude).
- Centrifuge at 16,000 × g (4°C, 20 minutes) to remove debris.
3.4 First Affinity Purification (Protein A/IgG Agarose)
Resin Equilibration: Use lysis buffer to pre-wash IgG agarose.
Binding: Incubate lysate with resin at 4°C for 2 hours (or overnight).
Wash Steps:
1) Base wash: 3 column volumes (CV) of lysis buffer.
2) High-salt wash: 3 CV lysis buffer + 500 mM NaCl.
3) Detergent wash: 3 CV lysis buffer + 0.5% sodium deoxycholate.
TEV Protease Cleavage: Elute complexes using TEV protease (1:50 enzyme:substrate ratio) in buffer containing 1 mM DTT.
3.5 Second Affinity Purification (CBP/Calmodulin Resin)
Resin Equilibration: Use calcium-containing buffer (1 mM CaCl₂).
Binding: Incubate TEV eluate with calmodulin resin at 4°C for 1 hour.
Wash and Elution:
- Base wash: 3 CV calcium buffer.
- Stringent wash: 3 CV calcium buffer + 500 mM NaCl.
- Elute with EGTA-containing buffer to disrupt calcium-dependent binding.
3.6 Sample Preparation for MS Analysis
- Concentration and Desalting: Use 10 kDa centrifugal filters (e.g., Amicon) and 50 mM NH₄HCO₃.
- Reduction/Alkylation: Treat with DTT (5 mM, 56°C, 30 min) and iodoacetamide (15 mM, RT, 30 min).
- Digestion: Trypsin (1:50 w/w) at 37°C for 16 hours.
- LC-MS/MS: Analyze peptides using a C18 column (75 μm × 25 cm) and Q Exactive HF-X mass spectrometer in data-dependent acquisition (DDA) mode.
4. Data Analysis and Validation of Protein Interactions
4.1 MS Data Interpretation
- Database Search: Use MaxQuant with species-specific UniProt databases.
- False Discovery Control: Apply reverse database search (≤1% FDR) and filter for ≥2 unique peptides across replicates.
4.2 Interaction Validation
Bioinformatics:
- STRING Database: Predict known interaction networks.
- Cytoscape + ClueGO: Visualize modules and perform GO/KEGG enrichment.
Experimental Validation:
- Co-IP: Confirm interactions using targetand partner-specific antibodies.
- Fluorescence Colocalization: Co-express mCherry (target) and GFP (partner) for confocal imaging.
- Surface Plasmon Resonance (SPR): Quantify binding affinity (KD).
5. TAP vs Co-IP vs Yeast Two-Hybrid vs BioID vs APEX
TAP excels in isolating native complexes with minimal contaminants, making it ideal for studying low-abundance or transient interactions. In contrast, Co-IP is faster but suffers from antibody cross-reactivity. Yeast two-hybrid systems are useful for high-throughput screens but lack physiological relevance. BioID and APEX are powerful for mapping proximal interactions but require careful optimization. The choice of technique depends on the biological question, sample type, and desired resolution.
TAP vs Co-IP vs Yeast Two-Hybrid vs BioID vs APEX
|
Technique |
Strengths |
Limitations |
Ideal Use Cases |
|
TAP |
High specificity, preserves native complexes |
Time-consuming, potential tag interference |
Low-abundance proteins, dynamic complexes |
|
Co-IP |
No genetic engineering, rapid |
High background, antibody-dependent |
Validating known interactions |
|
Yeast Two-Hybrid |
High-throughput, detects weak interactions |
High false positives, binary interactions |
Initial screening of interaction candidates |
|
BioID |
Proximity labeling, no purification needed |
Broad labeling radius (~10 nm) |
Transient/spatial interactions |
|
APEX/HRP |
Spatiotemporal resolution |
Requires enzyme overexpression |
Subcellular-specific interactions |
6. Innovations and Future Perspectives in TAP
Recent advancements are enhancing TAP’s utility:
- Cryo-EM Integration: Combining TAP-purified complexes with cryo-EM for high-resolution structural insights.
- Single-Step TAP: Developing smaller, non-disruptive tags (e.g., HaloTag) to streamline workflows.
- Quantitative MS: Using SILAC or TMT labeling for stoichiometric analysis of complex components.
Tandem Affinity Purification remains a cornerstone technique for dissecting protein interaction networks. Its compatibility with downstream analyses like MS and structural biology ensures its continued relevance in the era of multi-omics. By understanding its principles and limitations, researchers can leverage TAP to uncover novel biological mechanisms and therapeutic targets.


