Spatial Metabolomics: Transforming Biomedical and Agricultural Research
In recent years, spatial metabolomics has emerged as a game-changer in both medical and agricultural research. Unlike traditional bulk metabolomics, which offers a broad overview of metabolic activity, spatial metabolomics provides a precise, location-specific map of metabolite distribution within tissues. This unparalleled level of detail opens new possibilities for understanding complex biological processes, diagnosing diseases, and optimizing agricultural practices.
Do you know about spatial metabolomics? Why is this technology gaining so much attention, and what makes it so powerful? Does your research need its ability to uncover metabolic secrets hidden within tissues? Let’s unravel the mysteries of spatial metabolomics and explore the workflow of spatial metabolomics to discover how this cutting-edge tool can enhance your research.
Spatial Metabolomics-A Cutting-Edge Frontier in Biotechnology
On June 27th 2023, the World Economic Forum announced its annual list of breakthrough technologies, highlighting the most promising innovations with the potential to positively impact the world. Among these technologies, Spatial Omics was included as one of the top 10 breakthrough technologies. According to the Forum's report, the market for spatial omics is projected to reach $587 million by 2030, signaling rapid growth in this transformative field.
Spatial omics is rapidly emerging as a significant area of focus in biotechnology, following the single-cell sequencing technologies. It offers a novel approach to studying the spatial context of biological molecules within tissues, enabling a deeper understanding of complex biological systems. This technology encompasses various subfields, including spatial metabolomics, spatial proteomics, and spatial transcriptomics, each contributing to a more nuanced view of cellular function and disease mechanisms.
Spatial metabolomics, a key subset of spatial omics, has become a critical tool in modern medical research. By mapping the distribution of metabolites within tissues, spatial metabolomics provides unprecedented insights into how metabolic processes are organized at the cellular and tissue levels. This capability is opening new avenues for understanding diseases, optimizing drug development, and advancing precision medicine. A search on PubMed (1998-2022) using keywords like "Mass Spectrometry Imaging," "Spatial Metabolomics," or "Spatially Resolved Metabolomics" reveals a steady increase in the number of publications on spatial metabolomics over the years. MSI (Mass Spectrometry Imaging) continues to be one of the most prominent and rapidly evolving research areas, with significant growth in spatial metabolomics findings.
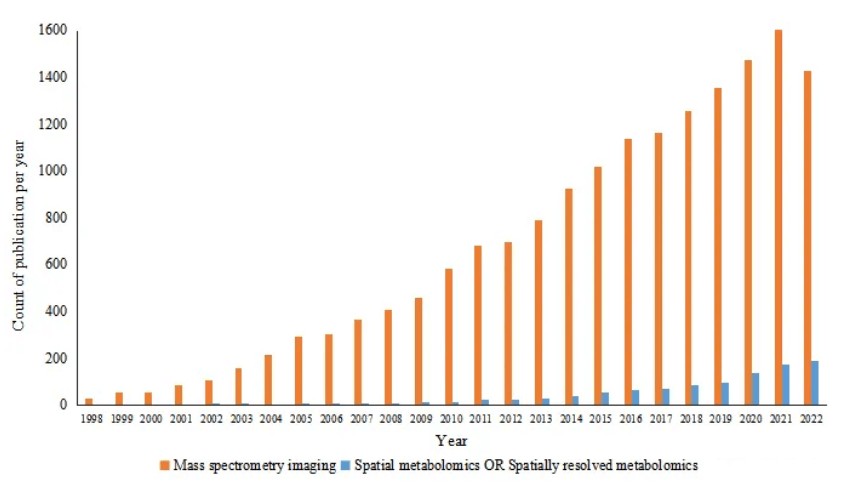
Number of publications related to spatial metabolomics in recent 20 years
How Spatial Metabolomics is Revolutionizing Metabolite Profiling- In Situ Visualization
Metabolomics enables the analysis of all small molecule metabolites in an organism or tissue and allows for the study of their dynamic changes, detecting short-term variations and providing terminal information about the biological system. It serves as an omics technology that offers a "real-time report" of the organism's current state. Traditional metabolomics techniques, such as targeted or untargeted liquid chromatography-mass spectrometry (LC-MS), provide valuable insights into the overall metabolic profile of a sample. However, these methods typically analyze homogenized tissue samples, offering only a bulk measurement of metabolites without spatial context. This limitation can mask important spatial variations that are crucial for understanding complex biological processes.
In contrast, spatial metabolomics focuses on the precise measurement of metabolite types, concentrations, and spatial distributions within tissues. Unlike traditional metabolomics, which provides bulk data, advanced spatial metabolomics incorporates Mass Spectrometry Imaging (MSI) technology to visualize metabolites in situ. MSI was first introduced in the late 20th century, initially for localizing proteins in tissue sections, before its focus shifted to detecting metabolites, lipids, and small molecules. The concept of spatially resolved metabolomics was proposed in 2007, primarily using laser microdissection techniques. After the introduction of MSI in 2010, the detection process became faster and more efficient. Over the past two decades, MSI has become one of the fastest-growing areas of mass spectrometry, with various ionization methods being employed, including MALDI, SIMS, DESI, and LAESI.
_1735624129_WNo_1179d313.jpg)
MS-based technologies currently used for metabolomics (Ren et al., 2018)
Before MSI, spatial information on biological processes in tissues was typically obtained through histological staining or immunohistochemistry. However, histological staining lacks molecular specificity, and immunohistochemistry requires prior knowledge of the target analyte, limiting the number of analytes that can be studied at any given time. MSI, based on mass spectrometry, directly scans biological samples, dividing the sample surface (usually tissue sections) into a virtual pixel grid. A mass spectrum is then generated for each pixel, representing the relative intensity of molecules in that region. Coupled with specialized image processing software, MSI enables high-throughput imaging of multiple substances without the need for isotopic labeling or staining, offering a more comprehensive and efficient analysis.
 dataset represents a collection of spectra acquired from a raster of _1735624152_WNo_1200d347.jpg)
An imaging mass spectrometry (MS) dataset represents a collection of spectra acquired from a raster of pixels (Alexandrov, 2020)
The Workflow of Spatial Metabolomics: From Sample Preparation to Data Analysis
Compared to bulk metabolomics, the key advantage of Mass Spectrometry Imaging (MSI) is its ability to perform in situ analysis of metabolic profiles. The experimental procedure typically involves several key steps:
1) Sample Preparation:
The first step in spatial metabolomics involves proper sample collection and preparation. Biological tissues, such as plant or animal tissues, are typically frozen or fixed to preserve their metabolic state and prevent degradation. The samples are then sectioned into thin slices (usually around 10-20 µm thick) using a cryostat or microtome. These tissue sections are placed onto a suitable substrate, such as a glass slide or a conductive film, depending on the imaging technique to be used.
2) Mass Spectrometry Imaging (MSI):
MSI techniques, such as Matrix-Assisted Laser Desorption/Ionization (MALDI) and Desorption Electrospray Ionization (DESI), are used to perform high-resolution, in situ analysis of metabolites.
- MALDI-MSI involves applying a matrix to the tissue section, followed by laser irradiation to desorb and ionize metabolites. The mass spectrum of each ion is recorded at multiple locations across the sample, generating a spatially resolved map of metabolites.
- DESI-MSI involves direct ionization of metabolites from the tissue surface using a charged spray of solvent, without the need for matrix application. This method allows for rapid, high-throughput imaging.
3) Data Acquisition:
The mass spectrometer scans the tissue sample pixel by pixel, capturing mass spectra at each point. The result is a set of data representing the intensity of ionized metabolites at each spatial coordinate. Advanced mass spectrometers with high resolution and sensitivity are employed to obtain detailed and accurate spectra, which are then correlated with specific regions of the tissue.
The data analysis in MSI includes the following steps:
1) Signal Processing: The first step involves saving the mass spectrometry data obtained from each pixel. Software is then used to perform signal processing tasks such as spectrum normalization, peak detection, baseline correction, and peak alignment.
2) Visualization of MSI Data: MSI provides intuitive and easy-to-interpret graphical results, showing which metabolites have been detected, where they are located within the tissue, and identifying spatial relationships between different molecules.
3) Quantification of Metabolites: For the obtained data, metabolite signals can be processed based on H&E (Hematoxylin and Eosin) staining results or through clustering techniques such as t-SNE or UMAP. These approaches enable the partitioning of metabolites into regions, and quantification of the metabolite signals in these regions, yielding quantitative data.
4) Qualitative Identification of Metabolites: By integrating databases, metabolites are qualitatively identified, including the annotation of molecular formulas and other relevant information.
5) Data Analysis: Further analyses are performed on the metabolite data, including differential analysis between different regions, multivariate statistical analysis across different samples (such as PCA, PLS-DA, or OPLS-DA), differential KEGG enrichment analysis, and ROC (Receiver Operating Characteristic) analysis. These analyses help uncover the differential distribution of metabolites across regions or samples, their biological significance, and their potential as biomarkers.
_1735624183_WNo_1200d336.jpg)
Steps of a typical data analysis workflow in imaging mass spectrometry (Alexandrov, 2020)
Key Applications of Spatial Metabolomics in Biomedical and Agricultural Research
Spatial metabolomics is rapidly emerging as a powerful tool in both biomedical and agricultural research. By providing detailed, spatially resolved metabolic profiles, it allows researchers to explore complex biological systems with a level of precision that traditional metabolomics cannot match. In biomedical research, spatial metabolomics is helping to uncover disease mechanisms, identify biomarkers, and guide drug development. In agriculture, it is enhancing our understanding of plant metabolism, development, stress responses, and crop improvement. Below, we explore key applications of spatial metabolomics in these two research fields.
Physiological Research:
A study published in Science Advances in March 2020 first discovered that gut microbiota produce carnitine-like analogs, which serve as communication mediators in the gut-brain axis. The researchers used MALDI-MSI and DESI-MSI to identify metabolites present in specific-pathogen-free (SPF) C57BL/6 mice but absent in germ-free (GF) mice. One molecule (m/z = 160.133) showed more than a 20-fold difference in expression between the two groups (Figure 5). NMR analysis revealed that this molecule consists of two isomers, and bacterial culture studies confirmed that it was produced by anaerobic symbiotic bacteria of the Lactobacillaceae family. The metabolite co-localized with carnitine in white matter and was found to inhibit carnitine-mediated fatty acid oxidation.
MALDI-MSI on brain and gut sections from C57BL/6 GF and SPF mice (Hulme et al., 2020)
Cancer Research:
A study published in Acta Pharmaceutica Sinica B in November 2019 investigated the inhibitory effect of 3-O-acetyl-11-keto-β-boswellic acid (AKBA) on glioma. Using MSI technology, the researchers analyzed the content and distribution of substances in the brain of an in situ mouse model. They found that AKBA improved the abnormal concentrations of phospholipids, fatty acids, amino acids, antioxidants, and other small molecules in glioblastoma xenografts, while reducing the abnormal accumulation of glucose in glioma. AKBA also inhibited autophagy by modulating the ERK and P53 signaling pathways.
MALDI-MSI of glioma tissues (Li et al., 2020)
Plant Toxicology:
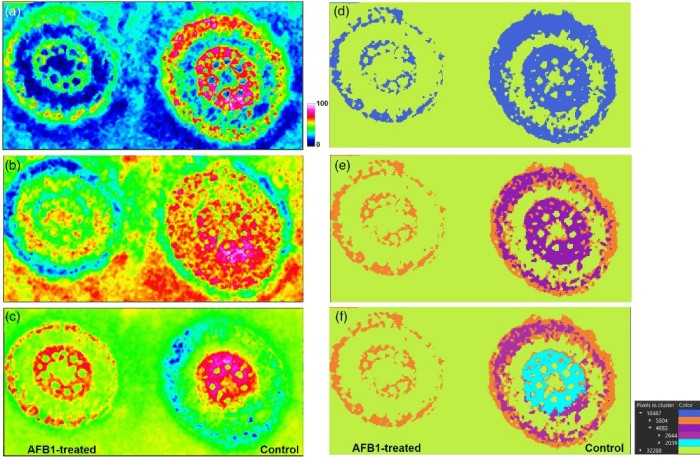
An article published in The Plant Journal in January 2021 revealed the interactions between maize and aflatoxin B1 (AFB1) at the tissue and organ level. To cope with harmful compounds, plants can biotransform xenobiotics, transfer parent compounds and metabolites, and compartmentalize them at the cellular or tissue level. This is also true for fungal toxins, such as the secondary metabolites of fungi that play a prominent role in plant infections. The study used AP-SMALDI MSI with a 5μm resolution to investigate the biotransformation, localization, and subsequent effects of AFB1 on primary and secondary metabolism in maize. It was found that AFB1 localized to the intercellular spaces of the roots and co-localized with the Phase I metabolite aflatoxin M2. Additionally, untargeted metabolomics revealed the greatest impacts on the biosynthesis of anthocyanins in the roots and chlorophyll metabolism, both of which were inhibited.
Exploratory data analysis for AP-SMALDI imaging of a maize root sample to visualize inter- and intra-sample comparison (Righetti et al., 2021)
Fruit Development:
An article published in Talanta in July 2021 studied the distribution and changes of endogenous molecules in Lycium barbarum (goji berry) fruits at three developmental stages. The results revealed that metabolites like choline, betaine, and citric acid were evenly distributed throughout the fruit at all stages of development. Hexose was found in the inner fruit peel and flesh, while sucrose was concentrated in the seeds. During fruit development, several phenolic acids and flavonoids accumulated in the outer peel, suggesting a protective role during biotic and abiotic stress in the growth process of the fruit. In terms of content, citric acid decreased with developmental signals, while choline, betaine, hexose, and sucrose increased.
MALDI-MSI was performed to wolfberry fruit at different development stages (Zhao et al., 2021)
Reference:
1. Alexandrov T. (2020). Spatial Metabolomics and Imaging Mass Spectrometry in the Age of Artificial Intelligence. Annual review of biomedical data science, 3, 61–87. https://doi.org/10.1146/annurev-biodatasci-011420-031537
2. Ren, J. L., Zhang, A. H., Kong, L., & Wang, X. J. (2018). Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC advances, 8(40), 22335–22350. https://doi.org/10.1039/c8ra01574k
3. Hulme, H., Meikle, L. M., Strittmatter, N., van der Hooft, J. J. J., Swales, J., Bragg, R. A., Villar, V. H., Ormsby, M. J., Barnes, S., Brown, S. L., et al., (2020). Microbiome-derived carnitine mimics as previously unknown mediators of gut-brain axis communication. Science advances, 6(11), eaax6328. https://doi.org/10.1126/sciadv.aax6328
4. Li, W., Ren, L., Zheng, X., Liu, J., Wang, J., Ji, T., & Du, G. (2020). 3-O-Acetyl-11-keto- β -boswellic acid ameliorated aberrant metabolic landscape and inhibited autophagy in glioblastoma. Acta pharmaceutica Sinica. B, 10(2), 301–312. https://doi.org/10.1016/j.apsb.2019.12.012
5. Righetti, L., Bhandari, D. R., Rolli, E., Tortorella, S., Bruni, R., Dall'Asta, C., & Spengler, B. (2021). Unveiling the spatial distribution of aflatoxin B1 and plant defense metabolites in maize using AP-SMALDI mass spectrometry imaging. The Plant journal : for cell and molecular biology, 106(1), 185–199. https://doi.org/10.1111/tpj.15158
6. Zhao, W. H., Zhang, Y. D., & Shi, Y. P. (2021). Visualizing the spatial distribution of endogenous molecules in wolfberry fruit at different development stages by matrix-assisted laser desorption/ionization mass spectrometry imaging. Talanta, 234, 122687. https://doi.org/10.1016/j.talanta.2021.122687
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


_1735624224_WNo_440d423.jpg)
_1735624259_WNo_706d672.jpg)
_1735624322_WNo_700d378.jpg)
