High-Sensitivity and High-Coverage Spatial Metabolomics
Spatial metabolomics is a scientific field focused on the in situ detection and annotation of metabolites within their spatial context. Recent studies highlight growing interest in metabolomics due to its critical role in elucidating metabolic regulation of molecular and cellular processes, the significance of non-genetic metabolic factors in maintaining tissue homeostasis, and its clinical applications. For instance, in oncology, spatial metabolomics aids in understanding tumor microenvironment dynamics, immune cell metabolism in metabolic/immune disorders, and cancer immunotherapy. These advancements have positioned spatial metabolomics as a pivotal technology. By integrating high-resolution analytical techniques with spatial information preservation, spatial metabolomics enables the analysis of metabolite types, concentrations, and distributions. It combines the chemical profiling capabilities of metabolomics with the spatial resolution of imaging technologies. The table below summarizes key distinctions among common mass spectrometry imaging (MSI) techniques.
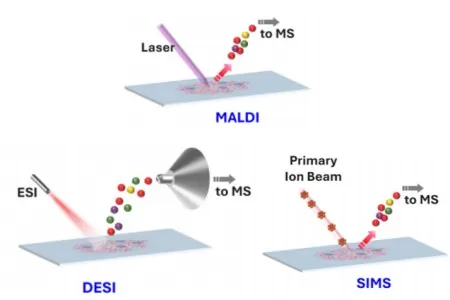
Common Mass Spectrometry Imaging Techniques
Table. Common mass spectrometry imaging (MSI) techniques
|
Technique |
Spatial Resolution |
Target Molecules |
Advantages |
Limitations |
|
MALDI-MSI |
5–100 μm |
Large/small molecules |
High sensitivity, broad applications |
Requires matrix coating, complex sample prep |
|
DESI-MSI |
50–200 μm |
Small molecules/lipids |
Matrix-free, suitable for live tissue |
Lower resolution, limited for large molecules |
|
SIMS |
100 nm–1 μm |
Small molecules |
Ultra-high resolution |
Poor ionization for peptides/large molecules |
Advantages of Spatial Metabolomics
(a) Spatial Information Preservation
Traditional metabolomics homogenizes tissue samples, losing spatial distribution data. In contrast, spatial metabolomics retains structural integrity while mapping metabolite localization.
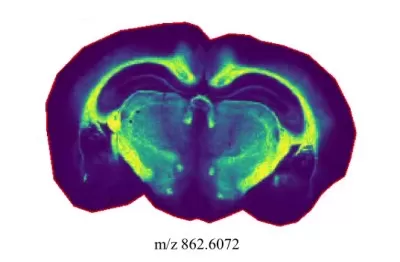
A spatial distribution map of one metabolite
(b) High-Resolution Imaging and Precise Localization
MSI achieves micrometer- or nanometer-level resolution, enabling single-cell metabolic studies and accurate spatial mapping of metabolites.
(c) High-Resolution Mass Spectrometry for Accurate Identification
Coupled with high-resolution mass spectrometry (HRMS), spatial metabolomics distinguishes metabolites with close m/z values, reducing false identifications.
(d) Multidimensional Data Integration
Beyond metabolite identification and quantification, spatial metabolomics integrates spatial distribution (e.g., staining maps) to reveal region-specific metabolic patterns.
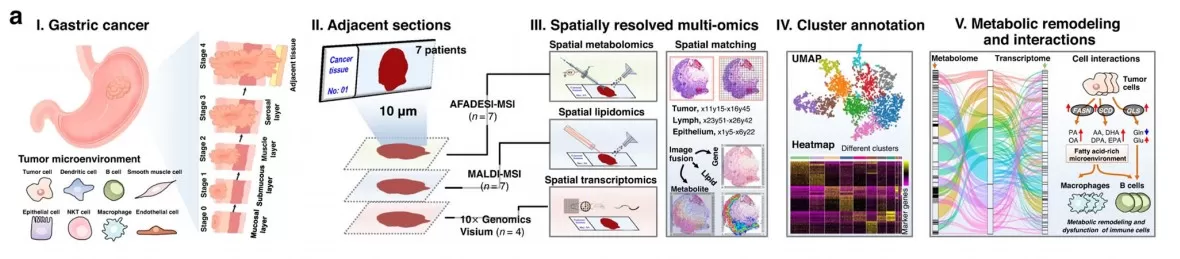
Spatial metabolomics data integration
(e) High Throughput and Multi-Omics Integration
Simultaneous detection of thousands of metabolites, combined with transcriptomic and proteomic data, supports comprehensive metabolic network modeling and systems biology.
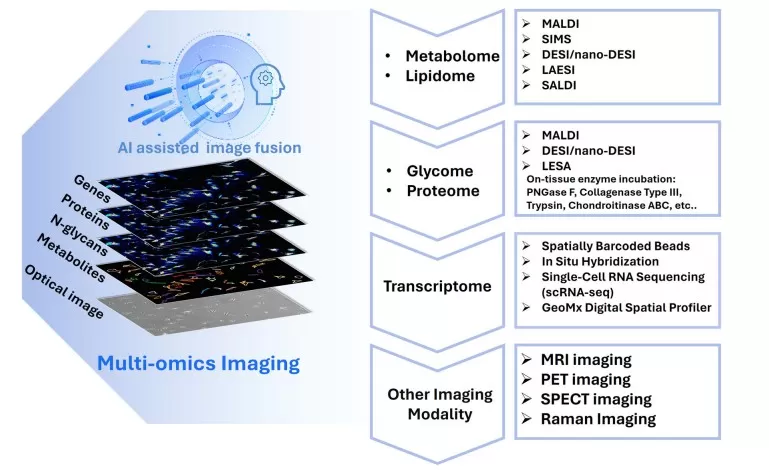
Spatial metabolomics for multi-omics integration
Technical Workflow
The workflow involves coating tissue sections with a UV-absorbing matrix to form co-crystals, followed by laser-induced ionization. A synchronized XY-stage moves the sample, enabling spatially resolved ion detection via mass spectrometry. Software processes raw spectra to generate metabolite distribution maps correlated with spatial coordinates.
Spatial Metabolomics Sample Preparation Guidelines
1. Sample Redundancy: Prepare≥2 replicates to account for potential quality issues.
2. Maximum Dimensions: 50 mm (length) × 30 mm (width) × 25 mm (thickness). Larger samples must be dissected fresh to avoid fracturing during freezing.
3. Minimum Dimensions: 1.5 mm × 1.5 mm × 2 mm. Smaller samples may require embedding or pre-sectioning.
4. Cryosectioning Considerations: Frozen sections are prone to ice crystal artifacts; test-sectioning is advised for structural integrity assessment.
5. Unembedded Samples: Protect against deformation or breakage during transport.
6. Embedding Protocol: Use one embedding block per sample to avoid cross-contamination.
References
Alexandrov T. Spatial metabolomics: from a niche field towards a driver of innovation. Nature Metabolism. 2023;5(9):1443-1445. doi:10.1038/s42255-023-00881-0.
Read more
- Spatial Metabolomics: Transforming Biomedical and Agricultural Research
- Spatial Metabolomics Explained: How It Works and Its Role in Cancer Research
- MALDI, DESI, or SIMS? How to Choose the Best MSI Techniques for Spatial Metabolomics
- How to Prepare Samples for Spatial Metabolomics: The Essential Guide You Need
- Unlocking Precision in Spatial Metabolomics: Essential Detection Parameters for Cutting-Edge Research


