Spatial Metabolomics in Cancer Research: Unlocking the Metabolic Code for Precision Oncology
Cancer metabolism has been a hot topic in oncology research for decades. However, the conventional approach to metabolomics has often failed to capture the intricate spatial variations of metabolic processes within tumor tissues. With the advent of spatial metabolomics, researchers can now visualize metabolic landscapes at a microscale resolution, unlocking new insights into tumor heterogeneity and metabolic reprogramming. This breakthrough is transforming our understanding of cancer biology and paving the way for more precise diagnostic and therapeutic strategies.
Why Spatial Metabolomics is Essential for Understanding Tumor Metabolism?
Traditional metabolomics has provided invaluable insights into cancer metabolism, but it largely treats tumors as homogeneous masses, averaging out regional differences. This approach overlooks crucial metabolic variations that occur within the tumor microenvironment (TME), where oxygen, nutrients, and cellular interactions vary significantly across different tumor regions. These differences can be profound, with metabolic activity in hypoxic tumor cores differing up to tenfold from well-vascularized outer regions.
Spatial metabolomics, through technologies such as mass spectrometry imaging (MSI), addresses these limitations by mapping metabolite distributions with spatial resolution. This approach enables researchers to:
- Precisely locate metabolic gradients, identifying how nutrients and metabolic waste accumulate in specific tumor regions.
- Correlate metabolic heterogeneity with gene expression and immune infiltration, providing a comprehensive view of tumor biology.
- Enhance clinical translation, enabling personalized medicine by identifying spatially distinct metabolic biomarkers.
By integrating metabolic information with spatial context, spatial metabolomics offers an unprecedented look into tumor biology, shaping the next generation of cancer diagnostics and therapies.
Five Key Applications of Spatial Metabolomics in Cancer Research
Spatial metabolomics bridges this gap by providing high-resolution maps of metabolite distributions in different tumor regions, offering deeper insights into tumor heterogeneity, metabolic interactions, and therapeutic vulnerabilities. By integrating spatial metabolomics with transcriptomics, proteomics, and imaging technologies, researchers have uncovered new metabolic biomarkers, treatment strategies, and mechanistic insights into tumor biology. Below, we explore five groundbreaking applications of spatial metabolomics in cancer research, demonstrating its potential to revolutionize cancer diagnostics and therapy.
1. Deciphering Metabolic Heterogeneity in the Tumor Microenvironment
The tumor microenvironment (TME) is a highly dynamic and heterogeneous ecosystem composed of cancer cells, immune cells, fibroblasts, and vasculature. Each of these components contributes uniquely to tumor growth, immune evasion, and drug resistance. However, their metabolic interactions have remained largely unexplored due to the limitations of traditional metabolomics techniques. Spatial metabolomics is now shedding light on these metabolic interactions by revealing how metabolites are distributed across different tumor regions. For example, using advanced mass spectrometry imaging, researchers have identified distinct metabolic signatures in hypoxic versus oxygen-rich tumor areas. The accumulation of glutamine and polyamines in specific regions of tumors, for instance, has been linked to aggressive cancer phenotypes.
Furthermore, studies combining spatial metabolomics with transcriptomics have shown how metabolic changes at the tumor-normal interface influence immune cell activity. Findings suggest that phosphatidylcholine (PC) metabolism suppression correlates with immune cell infiltration, revealing new potential targets for immunotherapy. By mapping these spatial metabolic variations, scientists can develop more effective therapeutic strategies that target specific metabolic vulnerabilities in tumors.
2. Mapping Tumor-Specific Metabolic Pathways to Unravel Cancer Progression
Cancer cells undergo profound metabolic reprogramming to support their rapid proliferation and survival. Unlike normal cells, which rely primarily on oxidative phosphorylation, cancer cells frequently adopt glycolysis (even in the presence of oxygen), a phenomenon known as the Warburg effect. However, this metabolic shift is not uniform across all cancer cells within a tumor. Spatial metabolomics allows researchers to track these metabolic changes at a highly localized level. For instance, a recent study published in Science identified that the oncometabolite D-2-hydroxyglutarate (D-2HG) inhibits CD8+ T-cell activity, weakening the immune system’s ability to fight cancer. This discovery has profound implications for cancer immunotherapy, as targeting D-2HG metabolism could restore immune function.
Similarly, researchers at Stanford University have used spatial metabolomics to map MYC-induced lipidomic changes in tumors, revealing a precise spatial distribution of lipid metabolism alterations. This level of insight into metabolic reprogramming not only enhances our understanding of tumor biology but also opens the door for new metabolic-targeted therapies.
3. Identifying Spatially Resolved Biomarkers for Cancer Diagnosis and Prognosis
Early and accurate cancer detection remains a major challenge in oncology. Biomarkers—molecular indicators of disease presence and progression—are crucial for diagnosing cancer at an early stage and predicting patient outcomes. However, traditional metabolomics often fails to distinguish between tumor-specific metabolic changes and systemic alterations occurring in non-cancerous tissues. Spatial metabolomics addresses this challenge by identifying metabolites that are uniquely expressed in tumor regions. For example, in clear cell renal cell carcinoma (ccRCC), researchers have discovered spatially distinct metabolic signatures linked to tumor progression, including elevated glycolysis and glutaminolysis. By correlating these metabolic alterations with genomic data, scientists have been able to classify tumors into subtypes with different prognostic outcomes.
Moreover, this approach has been instrumental in identifying new metabolic biomarkers for cancer detection. A study integrating spatial metabolomics with artificial intelligence has developed a highly accurate method for detecting early-stage lung cancer based on lipid metabolic signatures, demonstrating the clinical potential of this technology. These discoveries are paving the way for more precise and personalized cancer diagnostics.
4. Enhancing Cancer Treatment by Targeting Spatial Metabolic Adaptations
Despite significant advancements in cancer treatment, therapy resistance remains a major hurdle in oncology. Tumors often develop resistance to chemotherapy, immunotherapy, and targeted therapies by altering their metabolic pathways. Understanding these metabolic adaptations at a spatial level is crucial for developing more effective treatments. Spatial metabolomics has been instrumental in evaluating tumor responses to therapy. By mapping metabolic changes before and after treatment, researchers can identify metabolic pathways associated with drug resistance. For instance, in gastric cancer, studies have shown that cancer cells rely on metabolic crosstalk with surrounding fibroblasts to evade therapy. Targeting these metabolic interactions could lead to more effective combination therapies.
Additionally, spatial metabolomics is being used to explore new therapeutic targets. By identifying metabolic vulnerabilities unique to specific tumor regions, scientists are developing drugs that selectively target cancerous cells while sparing normal tissues, minimizing side effects and improving treatment efficacy.
5. In Situ Validation of Metabolic Signatures for Translational Oncology
A major challenge in cancer research is ensuring that metabolic findings from cell cultures and animal models are applicable to human tumors. Spatial metabolomics enables in situ validation of cancer-associated metabolites, confirming their biological relevance in human tissues. For example, in oral squamous cell carcinoma (OSCC), spatial metabolomics has revealed how metabolic reprogramming in epithelial-mesenchymal transition (EMT) regions drives tumor progression. By integrating this approach with spatial transcriptomics and immunohistochemistry, researchers can validate these findings at the molecular level. Such validation enhances the reliability of metabolic biomarkers and therapeutic targets, accelerating their translation into clinical applications.
Real-World Applications: Case Studies in Spatial Metabolomics
As spatial metabolomics continues to evolve, its applications in cancer research are demonstrating groundbreaking potential. By mapping the spatial distribution of metabolites in tumors, researchers are uncovering new diagnostic biomarkers, understanding metabolic interactions in the tumor microenvironment, and developing targeted treatment strategies. Below, we explore three compelling case studies where spatial metabolomics has made significant strides in advancing cancer research.
Case Study 1: Spatial Metabolomics Enables Early Lung Cancer Detection with AI Precision
Lung cancer remains one of the deadliest malignancies worldwide, with early detection being crucial for improving patient survival rates. However, existing screening methods, such as low-dose CT scans, have limitations in specificity and cost-effectiveness. A groundbreaking study published in Science Translational Medicine, leveraged spatial metabolomics, single-cell transcriptomics, and AI-driven lipidomics to develop an innovative approach for early lung cancer detection.
The study first utilized single-cell transcriptomic sequencing to analyze metabolic alterations in early-stage lung cancer tissues. The results revealed that lipid metabolism pathways were extensively dysregulated across different tumor cell types. Researchers then performed plasma lipidomic profiling on 171 early-stage lung cancer patients and 140 healthy individuals. Using mass spectrometry imaging (MSI) for spatial metabolomics, they confirmed that specific lipid metabolites were enriched in tumor tissues compared to normal lung tissue. Through advanced machine learning algorithms, the team identified nine lipid biomarkers that could reliably distinguish early-stage lung cancer patients from healthy individuals. These biomarkers were further validated in a cohort of over 2,100 clinical samples, achieving a diagnostic accuracy exceeding 90%. The sensitivity of this model was also over 90% in independent clinical cohorts, making it a promising tool for large-scale lung cancer screening.
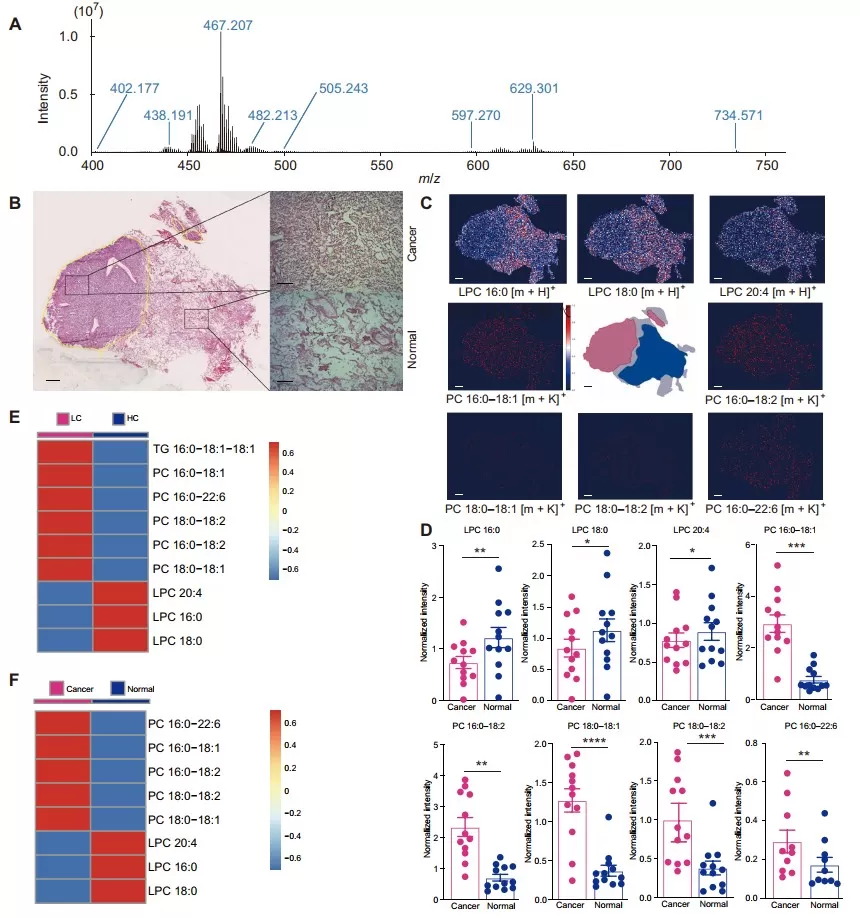
MALDI-MSI analyses of feature lipids of LCAID v2.0 in early-stage lung cancer tissues in situ (Wang et al., 2022)
Furthermore, spatial metabolomics was used to validate the presence and localization of these lipid biomarkers directly in lung cancer tissue, reinforcing their biological relevance. This study not only demonstrates the power of spatially-resolved metabolic profiling in early cancer detection but also highlights the potential of AI-assisted metabolomics in developing non-invasive, high-accuracy diagnostic methods.
Case Study 2: Spatial Metabolomics Unveils Metabolic Reprogramming in Betel Nut-Induced Oral Cancer
Oral squamous cell carcinoma (OSCC) is a highly aggressive malignancy often associated with betel nut chewing, a common risk factor in Southeast Asia. While the link between betel nut consumption and OSCC has been established, the precise metabolic mechanisms driving carcinogenesis remained unclear until spatial metabolomics was applied. A study published in Advanced Science used spatial transcriptomics and spatial metabolomics to investigate the tumor microenvironment (TME) of betel nut-induced OSCC. The researchers collected fresh tumor tissue samples from four OSCC patients diagnosed with oral submucous fibrosis (OSF), a precancerous condition linked to betel nut use. Spatially-resolved metabolomic and transcriptomic profiling provided a high-resolution map of metabolic alterations in different tumor regions.
Key findings from the study include:
- Identification of Partial Epithelial-Mesenchymal Transition (pEMT) Phenomena: Spatial transcriptomics revealed that OSCC cells at the invasive front underwent pEMT, acquiring fibroblast-like characteristics and contributing to collagen deposition in the TME.
- Cellular Crosstalk in the TME: Spatial metabolomics uncovered extensive metabolic interactions between epithelial cells, fibroblasts, and immune cells, revealing that fibroblast-driven metabolic alterations play a key role in promoting OSCC progression.
- Metabolic Reprogramming in OSCC Development: Betel nut-induced OSCC exhibited significant metabolic shifts, particularly in arginine metabolism, glutaminolysis, and lipid biosynthesis, supporting tumor growth and immune evasion.
 Schematic diagram exhibiting the detection and analysis of Spatial Transcriptomics and Sp_1742786733_WNo_874d450.webp)
A) Schematic diagram exhibiting the detection and analysis of Spatial Transcriptomics and Spatial Metabolomics in the OSF‐derived OSCC. B) Unsupervised clustering analysis (UMAP) in Spatial Metabolomics (Zhi et al., 2024)
By leveraging spatial metabolomics, this study provided the first spatially-resolved metabolic landscape of OSCC, offering valuable insights into how metabolic alterations drive betel nut-induced carcinogenesis. These findings suggest that targeting specific metabolic pathways could serve as a novel therapeutic strategy for OSCC prevention and treatment.
Case Study 3: Spatial Metabolomics Maps Metabolic Heterogeneity in Gastric Cancer
Gastric cancer is a highly heterogeneous disease with complex metabolic interactions between tumor cells and their surrounding microenvironment. Recent advances in spatial multi-omics have provided new insights into metabolic remodeling in gastric cancer, leading to the discovery of potential therapeutic targets. A landmark study, published in Nature Communications, applied spatial metabolomics, lipidomics, and transcriptomics to generate the first comprehensive spatially-resolved metabolic map of human gastric cancer tissues.
Key discoveries from the study include:
- Distinct Metabolic Heterogeneity Across Tumor Regions: By integrating spatial metabolomics with transcriptomic data, the team identified significant metabolic variations between different tumor compartments. Tumor cores exhibited increased lipid synthesis and amino acid metabolism, while tumor margins showed a metabolic profile more supportive of immune cell infiltration.
- Metabolic Crosstalk Between Tumor and Immune Cells: The study revealed that tumor cells at the tumor-normal interface engaged in active metabolic interactions with immune cells, particularly in arginine and proline metabolism. These metabolic changes suppressed anti-tumor immune responses, allowing tumors to evade immune surveillance.
- Potential Therapeutic Targets: Based on these findings, researchers proposed that targeting arginine metabolism and lipid biosynthesis pathways could serve as effective therapeutic strategies for gastric cancer.
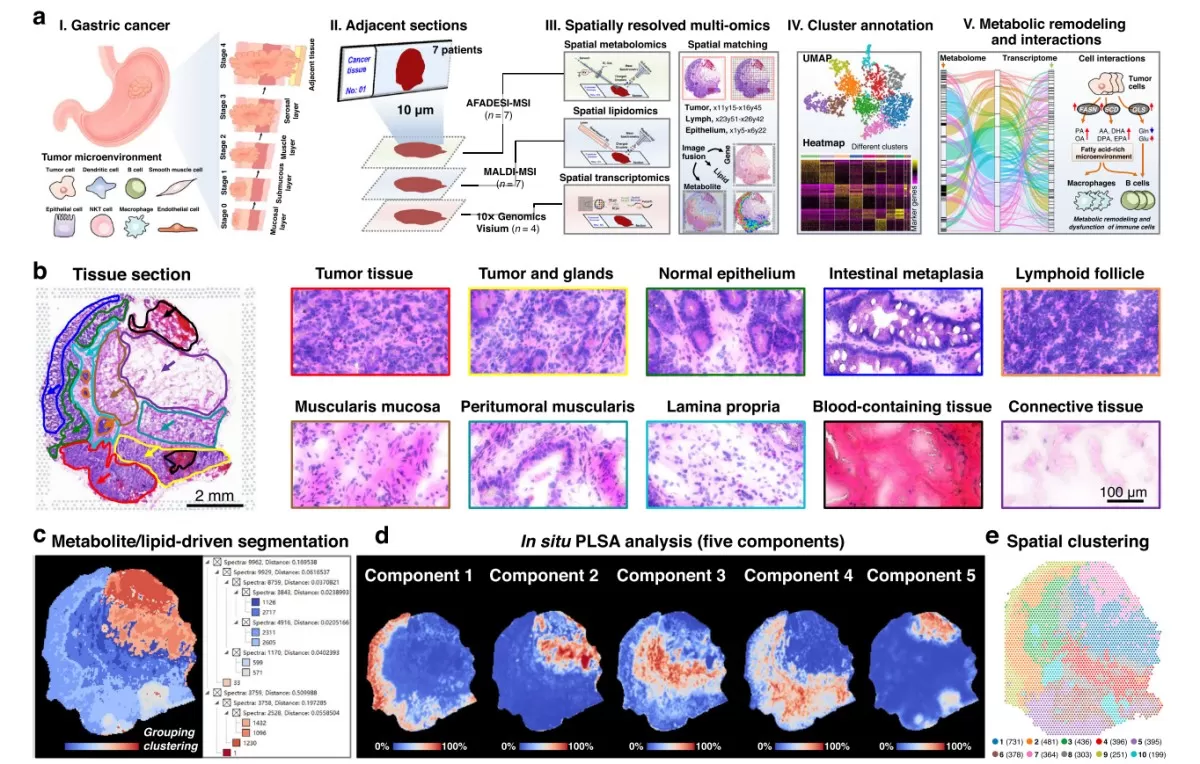
Spatially resolved multi-omics reveals intratumor heterogeneity of gastric cancer (Sun et al., 2023)
This study exemplifies how spatially-resolved multi-omics can uncover novel insights into tumor biology, paving the way for metabolic precision oncology. By mapping metabolic interactions at the cellular level, spatial metabolomics is driving the development of more effective, targeted therapies for gastric cancer.
The Future of Spatial Metabolomics in Personalized Cancer Therapy
The emergence of spatial metabolomics is revolutionizing cancer research, providing an unprecedented ability to map metabolic alterations with spatial precision. By integrating metabolomics with transcriptomics, proteomics, and imaging technologies, researchers are uncovering new layers of complexity in tumor biology.
As this field continues to evolve, its applications in precision oncology are expected to expand. The ability to identify tumor-specific metabolic vulnerabilities, track disease progression, and optimize therapeutic interventions holds immense promise for improving cancer outcomes. With ongoing advancements in mass spectrometry imaging and artificial intelligence, spatial metabolomics is set to play a pivotal role in the future of cancer diagnosis, prognosis, and treatment.
The era of spatial metabolomics has arrived, offering a powerful new lens through which we can decode the metabolic complexities of cancer. As research progresses, this innovative approach will undoubtedly lead to more effective, personalized, and targeted cancer therapies, bringing us closer to a world where cancer is not only treatable but also preventable.
Reference
Wang G, Qiu M, Xing X, et al. Lung cancer scRNA-seq and lipidomics reveal aberrant lipid metabolism for early-stage diagnosis. Sci Transl Med. 2022;14(630):eabk2756. doi:10.1126/scitranslmed.abk2756
Zhi Y, Wang Q, Zi M, et al. Spatial Transcriptomic and Metabolomic Landscapes of Oral Submucous Fibrosis-Derived Oral Squamous Cell Carcinoma and its Tumor Microenvironment. Adv Sci (Weinh). 2024;11(12):e2306515. doi:10.1002/advs.202306515
Sun C, Wang A, Zhou Y, et al. Spatially resolved multi-omics highlights cell-specific metabolic remodeling and interactions in gastric cancer. Nat Commun. 2023;14(1):2692. Published 2023 May 10. doi:10.1038/s41467-023-38360-5


