SomaScan Proteomics Technology: Principles and Advantages
Proteomics is a vital research field in the life sciences, focusing on the identification and quantification of all proteins within an organism. It provides crucial insights into complex biological processes and disease mechanisms. The SomaScan proteomics technology, developed by Somalogic, leverages nucleic acid aptamers known as SOMAmer (Slow Off-rate Modified Aptamers) to enrich and detect proteins with high specificity and sensitivity. This innovative platform allows for the simultaneous measurement of up to 11,000 proteins in a single, ultra-small sample, enabling comprehensive proteome profiling with unprecedented depth and breadth. In this blog, let’s delve into the technology behind SomaScan, exploring its innovative principles, unique advantages, and how it’s reshaping the landscape of proteomics.
1. Principles of SomaScan Proteomics Technology
The core technology of Somalogic is the SomaScan platform, which utilizes the specific binding of SOMAmer molecules to proteins for quantitative analysis. SOMAmer, or "Slow Off-rate Modified Aptamer," is a short single-stranded DNA molecule distinguished by its unique base sequences, each tailored to bind a specific protein with high affinity. Each SOMAmer is also equipped with special modification groups that enhance its ability to selectively adsorb to target proteins.
- The binding affinity of SOMAmers is attributed to several factors, including:
- Complementary Shape: The structural fit between the SOMAmer and the target protein.
- Polar Interactions: The attraction between polar regions of the molecules.
- Hydrophobic Interactions: The tendency of non-polar regions to associate.
- Hydrogen Bonding: The formation of bonds between hydrogen and electronegative atoms.
- Electrostatic Interactions: The attraction between oppositely charged groups.
It is important to note that SOMAmers do not form covalent bonds with their target proteins. The basic structure of the SOMAmer-protein complex is illustrated as follows:
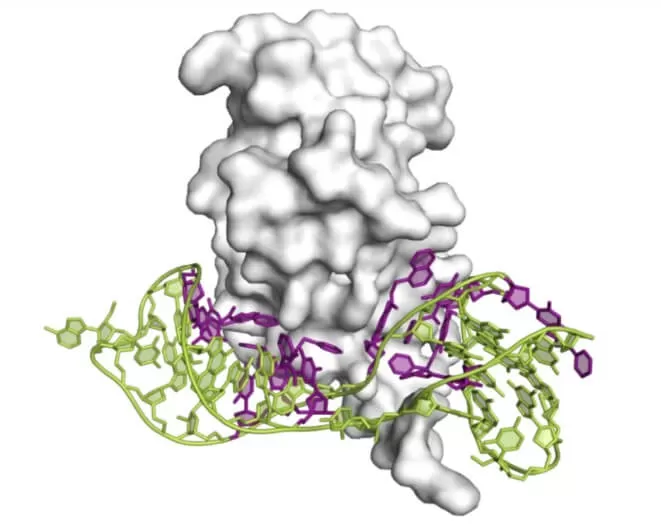 The pale section represents a schematic of the 3D structure of the IL-6 protein, while the green segment depicts the unmodified DNA strand. The purple portion illustrates the main structure of the SOMAmer DNA strand, which includes modification elements. The additional three groups consist of biotin, a photosensitive linker, and a fluorophore.
The pale section represents a schematic of the 3D structure of the IL-6 protein, while the green segment depicts the unmodified DNA strand. The purple portion illustrates the main structure of the SOMAmer DNA strand, which includes modification elements. The additional three groups consist of biotin, a photosensitive linker, and a fluorophore.
Biotin serves the purpose of forming a stable binding with streptavidin. The linker connects the biotin to the main structure of the SOMAmer. Notably, the linker is sensitive to ultraviolet light; upon exposure, it cleaves, effectively disconnecting the biotin from the SOMAmer core. The role of the fluorophore is crucial: when a SOMAmer labeled with the fluorophore binds to a chip, the intensity of the emitted fluorescence correlates with the quantity of the SOMAmer present. This, in turn, allows for the inference of the corresponding protein concentration, facilitating precise quantitative analysis in proteomics applications.
In summary, the core principles of SOMAmer technology can be encapsulated in three key points:
1. Specific Binding: SOMAmer molecules are meticulously designed to achieve highly specific binding with target proteins. Unlike antibodies, SOMAmers are based on nucleic acid sequences and, after chemical modifications, can attain binding affinity and specificity that equal or even surpass that of some conventional antibodies.
2. Chemical Modifications for Enhanced Stability: Chemical modifications to SOMAmer molecules enhance their stability in binding to target proteins and prolong the duration of this binding. This "slow off-rate" characteristic allows SOMAmers to function effectively under a broader range of biological conditions, thereby increasing detection sensitivity.
3. Fluorescent Labeling and Quantification: In the SomaScan platform, the binding of SOMAmer molecules to proteins is detected through fluorescence signals. The intensity of the fluorescence is directly proportional to the relative abundance of the protein, enabling quantitative analysis. This process allows for the simultaneous detection of thousands of proteins, providing comprehensive proteomic profiling.
2. Steps in the SomaScan Proteomics Detection Process
1. Modification of SOMAmer Reagents: Fluorophores, photolabile linkers, and biotin are covalently attached to the SOMAmer reagents (depicted in purple).
2. Capture of Target Proteins: The modified SOMAmer reagents are mixed with streptavidin beads, facilitating the capture of target proteins from biological samples.
3. Washing Away Unbound Proteins: Unbound proteins are thoroughly washed away, leaving only those that have specifically interacted with the biotinylated SOMAmer.
4. Photolytic Cleavage: Ultraviolet light is used to cleave the photolabile linker, effectively releasing non-specific complexes.
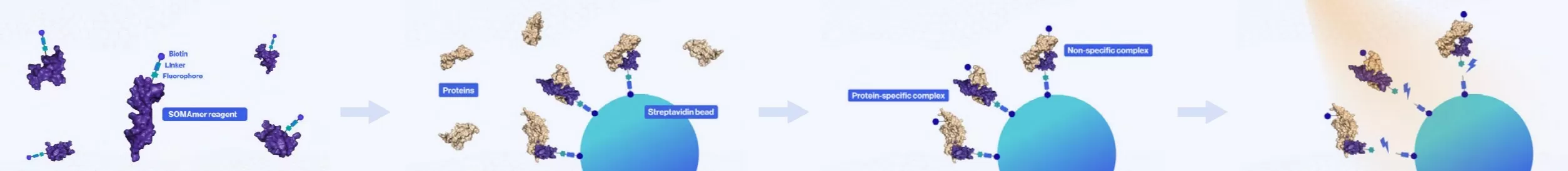
5. Dissociation of Non-Specific Complexes: Non-specific complexes are dissociated, while the specific complexes remain intact and bound.
6. Prevention of Rebinding: A polyanionic competitor (illustrated in green) is introduced to inhibit the re-association of any non-specific complexes.
7. Capture of SOMAmer-Biotinylated Protein Complexes: The SOMAmer-biotinylated protein complexes are captured by newly added streptavidin beads.
8. Release of SOMAmer from Complexes: The SOMAmer reagents are released from the complexes by denaturing the proteins. Following this, the SOMAmer hybridizes with complementary sequences on a microarray chip, where fluorescence detection is conducted. The fluorescence intensity detected on the microarray is directly proportional to the quantity of SOMAmer present in the original sample.

3. Advantages of SomaScan Proteomics Technology
Somalogic's SomaScan technology offers several significant advantages in the field of proteomics, particularly for large-scale analyses and clinical applications.
1. Ultra-High Throughput
The SomaScan platform can analyze over 11,000 proteins in a single experiment, greatly accelerating the processes of biomarker discovery, disease mechanism research, and drug screening. Compared to traditional proteomics techniques such as mass spectrometry, SomaScan's high-throughput capabilities make it especially well-suited for large-scale clinical sample screening.
2. High Sensitivity and Broad Dynamic Range
The sensitivity of the SomaScan platform is exceptionally high, allowing for the detection of low-abundance proteins, even at femtogram (10^-15 grams) concentrations. Additionally, SomaScan features an extensive dynamic range that spans ten orders of magnitude in protein abundance. This sensitivity and dynamic range make it ideal for analyzing diverse proteins in complex samples, such as blood and urine.
3. Minimal Sample Requirements
The SomaScan technology requires only a minimal volume of biological sample—approximately 50 microliters of plasma or serum—to perform comprehensive protein analyses. This is particularly important for studies involving precious samples, the discovery of biomarkers for rare diseases, and research with limited sample availability.
4. High Specificity and Precision
SOMAmer molecules exhibit a remarkable specificity for binding proteins, enabling them to effectively distinguish between structurally similar proteins. In comparison to traditional antibody-based detection methods, the SomaScan platform offers greater accuracy in protein identification and quantification, making it especially suitable for biomarker discovery, disease diagnosis, and monitoring.
5. Automation and Standardization
The SomaScan platform has achieved a high degree of automation and standardized processing, significantly reducing human error during operation. This enhances the reproducibility and reliability of the data, facilitating the technology's application in large-scale clinical studies and high-throughput screening in drug development.
Conclusion
Somalogic's SomaScan proteomics platform, with its unique SOMAmer molecules and high-throughput detection technology, has significantly advanced the field of proteomics research. Its advantages in sensitivity, throughput, automation, and precision highlight its vast potential applications in biomedical research, clinical diagnostics, and personalized medicine.
The SomaScan Assay currently includes services for detecting over 7,000 and 11,000 proteins, respectively, enabling high-throughput proteomic analysis and robust data interpretation. Looking ahead, as the technology continues to evolve, Somalogic's proteomics innovations will provide strong support for disease research, drug development, and precision medicine, facilitating groundbreaking discoveries in the life sciences.



