Single-Cell Proteomics: Mass Spec vs Single-Molecule Sequencing
This article delves into the exciting field of Single-Cell Proteomics and explores two powerful techniques: Mass Spectrometry and Single-Molecule Sequencing. We compare and contrast these methods, analyzing their strengths and limitations for uncovering the complex protein landscape within individual cells. Whether you're a researcher in proteomics, cell biology, or diagnostics, this guide provides valuable insights into the latest advancements and challenges in Single-Cell Proteomics, helping you choose the optimal approach for your research goals.
Table of contents
1. Introduction
1.1 What is Single-Cell Proteomics
1.2 How to perform single-cell proteomics
2.1 Introduction to Mass Spectrometry
2.2 Application of Mass Spectrometry in Single-Cell Proteomics
2.3 Advantages of Mass Spectrometry
2.4 Limitations of Mass Spectrometry
3.1 Introduction to Single-Molecule Sequencing
3.2 Application of Single-Molecule Sequencing in Single-Cell Proteomics
3.3 Advantages of Single-Molecule Sequencing
3.4 Limitations of Single-Molecule Sequencing
4. Discussion of Suitable Applications
5. Conclusion
1. Introduction
Single-cell proteomics is becoming a transformative tool in biological research and clinical applications. The ability to analyze proteins at the level of individual cells is providing new insights into cellular heterogeneity, disease mechanisms, and even potential drug targets. Traditional proteomics typically examines averaged data from a large number of cell samples, which can obscure the unique properties of individual cells. However, single-cell analysis allows scientists to capture the diversity and complexity of cellular functions, revealing how diseases develop and how to tailor treatments for maximum efficacy.
What is Single-Cell Proteomics
Single-cell proteomics is the study of proteins at the single-cell level, providing a higher-resolution approach to understanding complex biological systems. This approach is becoming increasingly popular in scientific research because it is able to reveal subtle differences between cells that were previously undetectable. By analyzing protein expression, modifications, and interactions in individual cells, single-cell proteomics has paved the way for advances in fields ranging from immunology to oncology.
How to perform single-cell proteomics
Currently, there are two main technologies at the forefront of single-cell proteomics: mass spectrometry and single-molecule sequencing. Each method has its own unique advantages and limitations, and understanding these differences is critical for researchers who want to apply single-cell proteomics in their own research.
Single-cell proteomics methods.
2. Mass Spectrometry
Introduction to Mass Spectrometry
Mass spectrometry (MS) is a powerful analytical technique that identifies and quantifies proteins based on their mass-to-charge ratio (m/z). In single-cell proteomics, MS enables researchers to detect and measure proteins with remarkable precision, even at trace levels. The basic process involves ionizing protein fragments (often peptides), accelerating them through an electric or magnetic field, and then detecting them based on their unique m/z values. Each protein generates a characteristic "fingerprint" that can be analyzed to determine its identity and abundance.
Application of Mass Spectrometry in Single-Cell Proteomics
In single-cell proteomics, mass spectrometry is employed to analyze the protein composition of individual cells. The process typically begins with the isolation of a single cell, followed by proteolysis, where enzymes break down the cell’s proteins into peptides. These peptides are then separated, often using liquid chromatography, before being introduced into the mass spectrometer. The mass spectrometer detects each peptide’s m/z value, allowing researchers to identify and quantify proteins unique to that cell.
Mass spectrometry has been instrumental in advancing single-cell proteomics because it provides an in-depth look at the protein composition within individual cells. By allowing researchers to identify and quantify thousands of proteins, MS offers insights into cell heterogeneity, protein expression profiles, and the dynamic changes associated with cellular processes and disease progression.
Advantages of Mass Spectrometry
High Sensitivity: Mass spectrometry can detect proteins present at extremely low levels, making it suitable for analyzing single-cell samples where protein quantities are minimal. High Resolution: MS offers high resolution, allowing it to distinguish between proteins of similar mass, which is crucial for studying complex cellular processes with precision. Wide Protein Coverage: Mass spectrometry can identify and quantify a large number of proteins, providing a comprehensive overview of a cell’s proteome. These strengths make mass spectrometry a robust tool for single-cell proteomics, as it can deliver detailed, high-quality data that would otherwise be difficult to obtain with other methods.
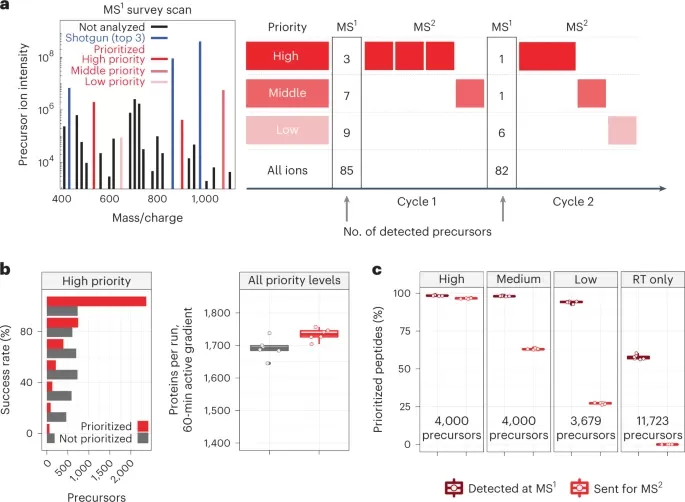
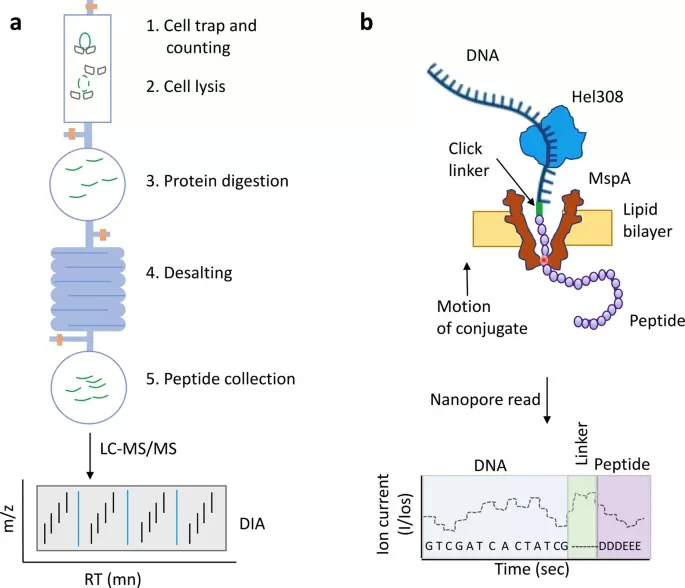
Mass spectrometry analysis of single-cell proteomes
Limitations of Mass Spectrometry
Low Throughput: Mass spectrometry can be time-consuming, especially when applied to single-cell samples. Analyzing each cell individually can extend experiment times significantly. High Cost: The equipment and reagents required for mass spectrometry are expensive, making this technique costly for large-scale studies or facilities with limited budgets. Requirement for Skilled Operators: Operating a mass spectrometer demands specialized training and expertise. Proper calibration, sample preparation, and data interpretation are essential, requiring experienced personnel. While mass spectrometry offers unparalleled insights into single-cell proteomics, these limitations highlight the challenges associated with using this technology on a routine basis. Researchers must weigh the benefits of high sensitivity and resolution against the logistical and financial costs, particularly when considering large-scale studies or clinical applications.
3. Single-Molecule Sequencing
Introduction to Single-Molecule Sequencing
Single-molecule sequencing (SMS) is an advanced technique that allows for the direct reading of protein sequences at the single-molecule level. Unlike mass spectrometry, which relies on analyzing fragmented peptides, single-molecule sequencing directly decodes the amino acid sequences of individual proteins. By observing each protein molecule individually, SMS provides a unique and detailed view of the protein’s structure, which can reveal important information about its function and modifications.
Application of Single-Molecule Sequencing in Single-Cell Proteomics
In single-cell proteomics, single-molecule sequencing offers a promising approach for analyzing protein expression in individual cells. Through direct sequencing, this technique can identify specific proteins and determine their abundance within a single cell, helping to map the unique protein profile of each cell. This direct sequencing process bypasses the need for extensive sample preparation, making it potentially faster and more efficient for high-throughput analysis of single-cell samples.
Single-molecule sequencing can provide valuable insights into protein expression patterns, particularly in heterogeneous cell populations. This is especially useful for understanding cellular diversity and identifying novel biomarkers in complex tissues, cancer, and other disease states.
Advantages of Single-Molecule Sequencing
-
High Throughput: Single-molecule sequencing can rapidly analyze a large number of single-cell samples, making it well-suited for studies that require high data volumes.
-
Direct Protein Sequence Reading: Unlike indirect methods, SMS directly decodes the amino acid sequences of proteins, offering comprehensive protein information, including post-translational modifications.
-
Lower Cost: Compared to mass spectrometry, single-molecule sequencing is generally more cost-effective, especially when dealing with large sample sizes or high-throughput studies.
These strengths make SMS a valuable tool in single-cell proteomics, especially when high-throughput and cost-effective analysis are essential.
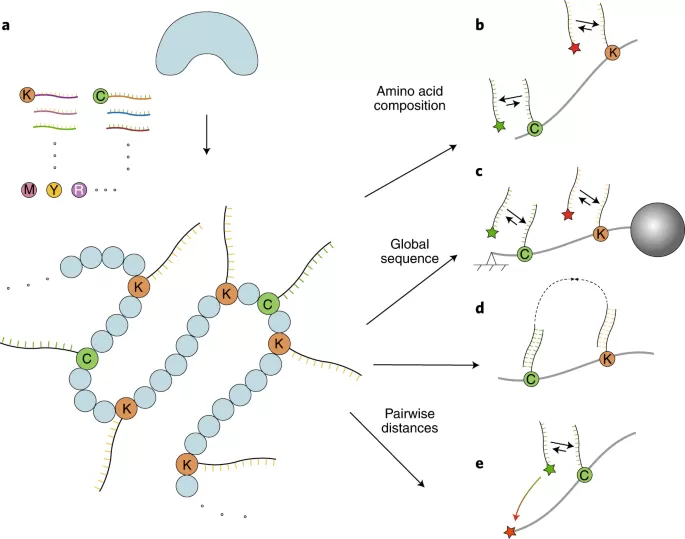
Single-molecule sequencing to analyze single-cell proteomes
Limitations of Single-Molecule Sequencing
-
Lower Sensitivity: Single-molecule sequencing generally requires higher protein concentrations to achieve accurate readings, which can limit its application to certain sample types.
-
Limited Coverage: Current single-molecule sequencing technologies can only sequence part of the protein’s sequence, restricting its ability to provide a complete proteomic profile.
-
Emerging Technology: Single-molecule sequencing is still rapidly evolving, and as a relatively new technology, it has yet to reach the maturity and robustness of mass spectrometry.
While single-molecule sequencing offers exciting possibilities in single-cell proteomics, these limitations mean that researchers must carefully consider the types of data they need and the resources available. As technology improves, single-molecule sequencing may become a more comprehensive solution for single-cell proteomics, bridging the gap between high-throughput sequencing and detailed protein analysis.
To provide a clear understanding of the differences between mass spectrometry and single-molecule sequencing in single-cell proteomics, the following table outlines the advantages and limitations of each technology:
| Feature | Mass Spectrometry (MS) | Single-Molecule Sequencing (SMS) |
|---|---|---|
| Sensitivity | High – can detect proteins at very low concentrations | Lower – requires higher protein concentration |
| Resolution | High – can distinguish proteins with similar masses | Moderate – provides partial protein sequence information |
| Throughput | Low – time-intensive for single-cell samples | High – can rapidly process large numbers of cells |
| Cost | Higher – requires expensive equipment and reagents | Lower – more cost-effective for high-throughput applications |
| Expertise Required | High – specialized training and expertise needed | Moderate – simpler workflow, although technology is evolving |
| Protein Coverage | Wide – can identify and quantify a broad range of proteins | Limited – partial sequence reading, restricts full profiling |
| Stage of Development | Mature and widely adopted in research labs | Emerging – rapidly developing but still less mature |
4. Discussion of Suitable Applications
Selecting the appropriate technology depends on several factors, including the study’s objectives, sample type, budget, and required data detail:
-
Mass Spectrometry (MS): This method is well-suited for studies that require in-depth proteomic profiling with high sensitivity and resolution. It is ideal when identifying a wide range of proteins is critical, such as in complex tissues or cancer research. However, for projects with limited budgets or needing rapid data collection, MS may be less practical due to its cost and lower throughput.
-
Single-Molecule Sequencing (SMS): SMS is an excellent choice for high-throughput applications where a large number of single cells need to be analyzed quickly and cost-effectively. It is particularly useful in studies where direct protein sequence reading is essential, though its current limitations in sensitivity and full-sequence coverage should be considered. This method may be more accessible for labs with budget constraints but still requires continued development to achieve broader protein coverage and improved sensitivity.
5. Conclusion
In single-cell proteomics, both mass spectrometry and single-molecule sequencing are invaluable technologies, each offering unique advantages and facing certain limitations. Mass spectrometry excels in sensitivity and resolution, providing a comprehensive view of the proteome, though it is often time-consuming and costly. Single-molecule sequencing, on the other hand, offers high-throughput and cost-effective analysis with direct sequence reading, but it currently faces challenges in sensitivity and protein coverage.
Selecting the most appropriate technology is crucial and depends on various factors, including the research objectives, sample types, budget, and the level of detail required. Researchers must carefully evaluate their needs to make the best choice between these methods, as each technique has the potential to significantly impact data quality and research outcomes.
6. Reference
1. Mansuri, M.S., Williams, K. & Nairn, A.C. Uncovering biology by single-cell proteomics. Commun Biol 6, 381 (2023). https://doi.org/10.1038/s42003-023-04635-2
2. Huffman, R.G., Leduc, A., Wichmann, C. et al. Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics. Nat Methods 20, 714–722 (2023). https://doi.org/10.1038/s41592-023-01830-1
3. Alfaro, J.A., Bohländer, P., Dai, M. et al. The emerging landscape of single-molecule protein sequencing technologies. Nat Methods 18, 604–617 (2021). https://doi.org/10.1038/s41592-021-01143-1
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.