Key Protein Extraction Techniques for Biological Fluids in Proteomics
Overview of Biological Fluids in Proteomics
Biological fluids refer to fluids associated with life phenomena, primarily including blood, saliva, exhaled breath condensate, urine, cerebrospinal fluid, aqueous humor, synovial fluid, cell culture media, fermentation broths, urine-like samples, lavage fluid, and dialysis fluid.
There is a wide variety of biological fluid samples, and significant differences exist among them. Some samples, such as urine, dialysis fluid, lavage fluid, and cell culture media, have large volumes but low protein concentrations. Others, like dialysis fluid and fermentation broths, may contain higher sugar levels. Some samples, such as saliva, cerebrospinal fluid, aqueous humor, synovial fluid, and tear fluid, have low protein content, small volumes, or are challenging to collect clinically. In contrast, samples like serum/plasma and milk have high protein content and are easier to obtain, but also contain abundant proteins. These sample types exhibit different requirements for protein extraction in proteomics, which can be categorized into three main groups.
Protein Extraction from Plasma/Serum and Milk Samples
Plasma and serum are commonly used clinical samples in proteomics research, particularly for identifying disease biomarkers and discovering new drug targets. However, these samples contain a large amount of high-abundance proteins, which complicates mass spectrometry (MS) analysis. Since MS is a saturation-based detection technique, signals from low-abundance proteins are often suppressed by high-abundance proteins, making their detection challenging. Increasing the depth of proteomic analysis in these samples by efficiently removing high-abundance proteins is a major focus in the field. Three mainstream approaches for high-abundance protein removal are:
 1. Non-Antibody-Based Methods: A prominent example is Bio-Rad's Proteominer kit. This kit selectively binds low-abundance proteins while excess high-abundance proteins, unbound to the matrix, are removed. Post-treatment, approximately 1,500 proteins can be identified using instruments like the timsTOF Pro2 in DIA mode. This method is species-agnostic, applicable to various sample types, cost-effective, and reproducible.
1. Non-Antibody-Based Methods: A prominent example is Bio-Rad's Proteominer kit. This kit selectively binds low-abundance proteins while excess high-abundance proteins, unbound to the matrix, are removed. Post-treatment, approximately 1,500 proteins can be identified using instruments like the timsTOF Pro2 in DIA mode. This method is species-agnostic, applicable to various sample types, cost-effective, and reproducible.
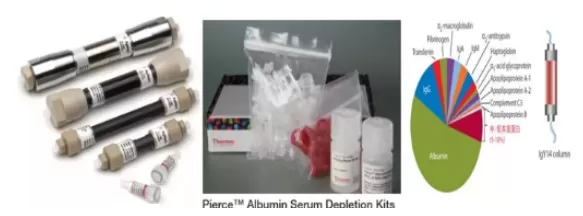
2. Antibody-Based Methods: Examples include Agilent's MARS columns, Thermo's Pierce Albumin/IgG kits, and SIGMA's Human IgY14 and SuperMix Columns. These products use specific antibodies to deplete high-abundance proteins in blood samples, significantly enhancing the depth of proteomic identification. These methods offer high stability and reproducibility but are expensive and have limitations in storage.
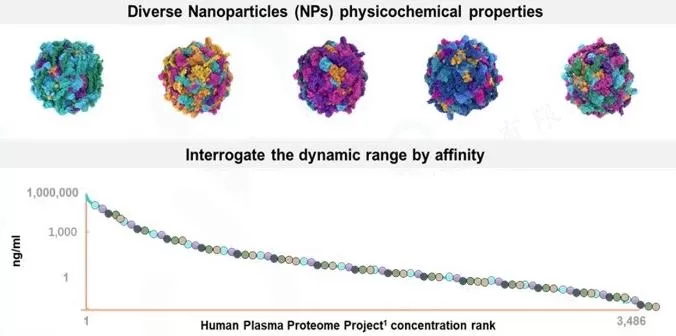
3. Nanomaterials: Seer’s Proteograph XT kit is a cutting-edge method based on nanomagnetic beads, which has transformed blood proteomics by enhancing identification depth by 2-3 times. The principle relies on the "protein corona" and Vroman effect to enrich low-abundance proteins. This technique combines the species-agnostic nature of Proteominer with the stability of antibody-based methods, offering deep, reproducible quantification of large-scale blood proteomes without the need for explicit removal of high-abundance proteins. Paired with advanced MS technology, this approach allows for highly accurate and reproducible large-scale blood proteomics analysis.
Protein Extraction from Urine, Fermentation Liquid, and Lavage Fluid
These samples are characterized by large volumes and low protein content, often containing various contaminants such as salts, sugars, and metabolites that can interfere with proteomic analysis. A common pre-processing technique for such samples is ultrafiltration concentration.
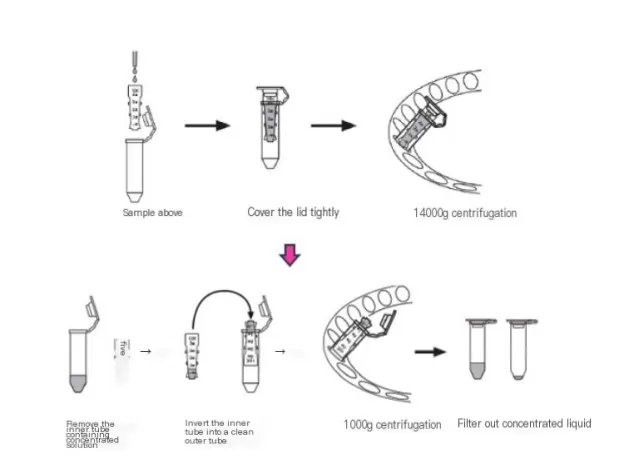 Ultrafiltration, using a 10kD filter (e.g., Millipore), is a key method for concentrating proteins in low-abundance samples. This technique increases protein concentration by filtering out unwanted components. After concentration, the sample solution is exchanged with 1x PBS buffer to remove interfering substances like pigments, sugars, and metabolites. The protein concentration is then measured using the BCA assay, preparing the sample for further proteomic analysis.
Ultrafiltration, using a 10kD filter (e.g., Millipore), is a key method for concentrating proteins in low-abundance samples. This technique increases protein concentration by filtering out unwanted components. After concentration, the sample solution is exchanged with 1x PBS buffer to remove interfering substances like pigments, sugars, and metabolites. The protein concentration is then measured using the BCA assay, preparing the sample for further proteomic analysis.
However, this method may result in the loss of low molecular weight proteins, and it is important to use fresh samples. Samples that have been stored for extended periods or subjected to multiple freeze-thaw cycles may suffer significant protein loss.
Protein Extraction from Saliva, Aqueous Humor, Cerebrospinal Fluid
These samples are typically small in volume with low protein content, making them challenging to collect in clinical settings. It's crucial to adhere strictly to sampling protocols to avoid complications during downstream processing.
For these types of samples, minimal pre-processing is often ideal. After collection, samples can be centrifuged at high speed, with the supernatant used for subsequent proteomic analysis. Before centrifugation, the sample’s condition post-thaw should be carefully evaluated. If the sample appears cloudy, it could be due to protein precipitation or the presence of lipids. Centrifugation can help distinguish whether the cloudiness is from lipid contamination or protein precipitation.
Overall, for biological fluid samples, it's essential to choose pre-processing methods based on the sample type. The general principle in proteomics is to maximize protein extraction while minimizing experimental errors and reducing the impact of the method on the sample itself.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


