Proteomics and Phosphoproteomics Uncover Novel Mechanisms
In the era of precision medicine, LC-MS-based proteomics and phosphoproteomics have emerged as indispensable tools for decoding complex biological systems. Both approaches leverage high-resolution LC-MS platforms for accurate identification and quantification, enable high-throughput profiling of biological samples, and hold critical translational relevance in biomarker discovery and therapeutic target identification. While both technologies rely on LC-MS to analyze proteins, they target distinct aspects of cellular machinery.
Proteomics vs. Phosphoproteomics: Key Similarities and Differences
|
Aspect |
Proteomics |
Phosphoproteomics |
|
Focus |
Global protein expression and abundance |
Site-specific phosphorylation modifications |
|
Target |
All detectable proteins in a sample |
Phosphorylated peptides/proteins only |
|
Enrichment Required |
No (unless subproteomes are targeted) |
Yes (e.g., TiO₂, antibodies, IMAC) |
|
Data Complexity |
High (thousands of proteins) |
Extremely high (dynamic PTM regulation) |
|
Biological Insight |
Protein expression changes |
Signaling pathway activation/regulation |
|
Technical Challenges |
Dynamic range, protein quantification |
Low stoichiometry, transient modifications |
Integrated Proteomics and Phosphoproteomics in Disease Research
Combining LC-MS-based proteomics and phosphoproteomics unlocks multi-dimensional insights into disease mechanisms. In cancer, proteomics identifies dysregulated proteins (e.g., HER2), while phosphoproteomics maps hyperactive kinase pathways (e.g., AKT/mTOR), revealing drivers of metastasis. For neurodegeneration, proteomics detects protein aggregates (e.g., tau), and phosphoproteomics highlights pathogenic phosphorylation (e.g., pTau in Alzheimer’s), linking accumulation to kinase dysregulation. In cardiovascular disease, proteomics tracks hypertrophy markers (e.g., SERCA2a), and phosphoproteomics uncovers calcium-handling defects (e.g., phospholamban phosphorylation), explaining maladaptive remodeling. Similarly, in metabolic disorders, proteomics profiles enzymes, while phosphoproteomics exposes insulin signaling flaws (e.g., IRS1 phosphorylation). This synergy bridges protein expression (“what”) with signaling dynamics (“how”), enabling precise biomarker discovery (e.g., phosphorylation-specific markers) and drug target identification (e.g., kinase inhibitors). By integrating these approaches, researchers decode complex pathologies and advance precision therapies.
Case study: How Does MYDGF Combat Pressure Overload-Induced Heart Failure?
As an example, here we are also excited to highlight a groundbreaking study titled "Myeloid-Derived Growth Factor Protects Against Pressure Overload-Induced Heart Failure by Preserving Sarco/Endoplasmic Reticulum Ca²⁺-ATPase Expression in Cardiomyocytes" published in Circulation. Leveraging conventional proteomics and phosphorylation-modified proteomics, the authors identified MYDGF as a key mediator of cardiac adaptation to chronic pressure overload. Using LC-MS for targeted protein quantification and phosphoproteomic profiling, the study revealed MYDGF’s role in enhancing SERCA2a expression via PIM1 kinase activation. Phosphoproteomics identified PIM1 as a key node, while conventional proteomics tracked SERCA2a expression, demonstrating the synergy between PTM (post-translational modification) and global protein analysis. This work underscores the power of multi-omics approaches in dissecting complex disease mechanisms and therapeutic opportunities.
Heart failure, a leading cause of morbidity and mortality worldwide, often develops in response to chronic hemodynamic stress such as hypertension or aortic stenosis. Persistent pressure overload triggers maladaptive cardiac remodeling, characterized by hypertrophy, fibrosis, and impaired calcium handling, ultimately leading to contractile dysfunction. While inflammation is traditionally viewed as detrimental in heart failure, recent studies suggest that certain immune-derived factors may confer protective effects. This study focuses on myeloid-derived growth factor (MYDGF), a protein secreted by monocytes and macrophages.
MYDGF Expression Rises in Pressure-Overloaded Hearts and Plasma
Using targeted LC-MS, the team measured MYDGF protein levels in murine and human samples. In mice subjected to transverse aortic constriction (TAC), MYDGF abundance surged in the left ventricle and plasma, peaking at day 3 and remaining elevated for 42 days. Similarly, patients with severe aortic stenosis showed elevated plasma MYDGF, which normalized after valve replacement. Proteomic profiling pinpointed monocytes and macrophages as the primary MYDGF sources, highlighting the immune system’s adaptive role in heart failure.
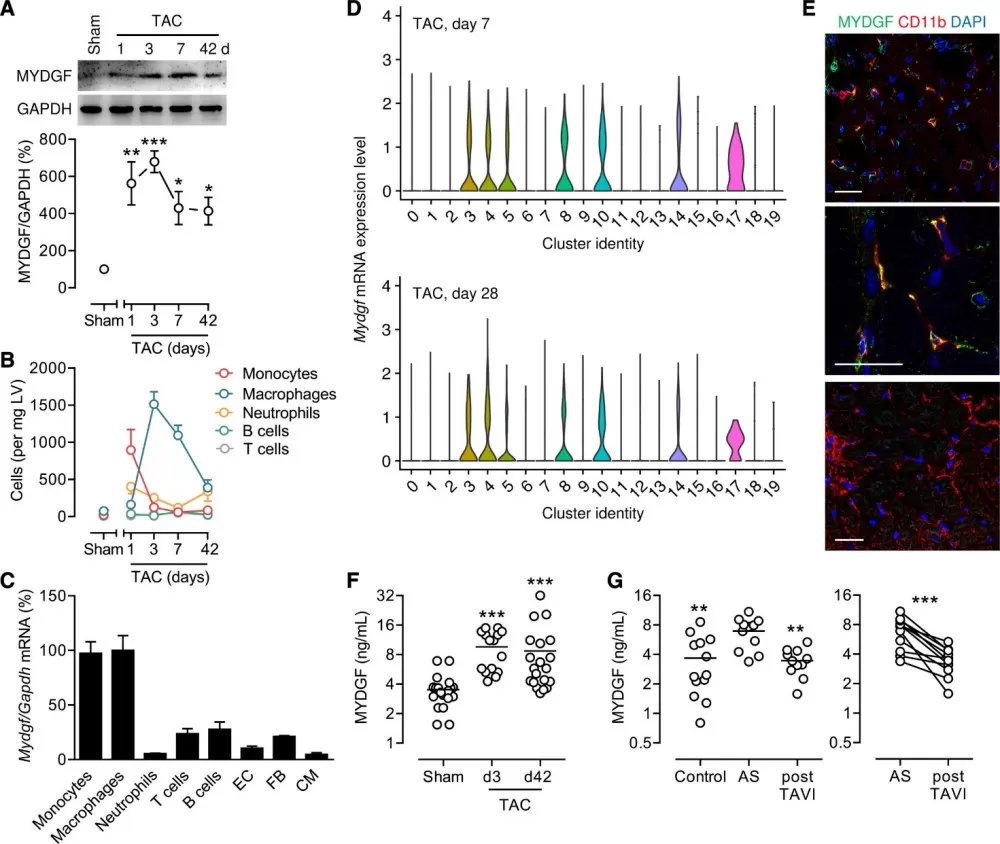
MYDGF expression during pressure overload
Phosphoproteomics Links MYDGF to PIM1-SERCA2a Signaling
To uncover MYDGF’s mechanism, phosphoproteomic analysis of neonatal rat cardiomyocytes revealed that MYDGF counteracted endothelin-1-induced hypertrophy by activating PIM1 kinase. Computational substrate-based kinase inference and siRNA experiments confirmed that MYDGF enhances PIM1 activity, which in turn upregulates SERCA2a—a calcium pump critical for cardiomyocyte function.
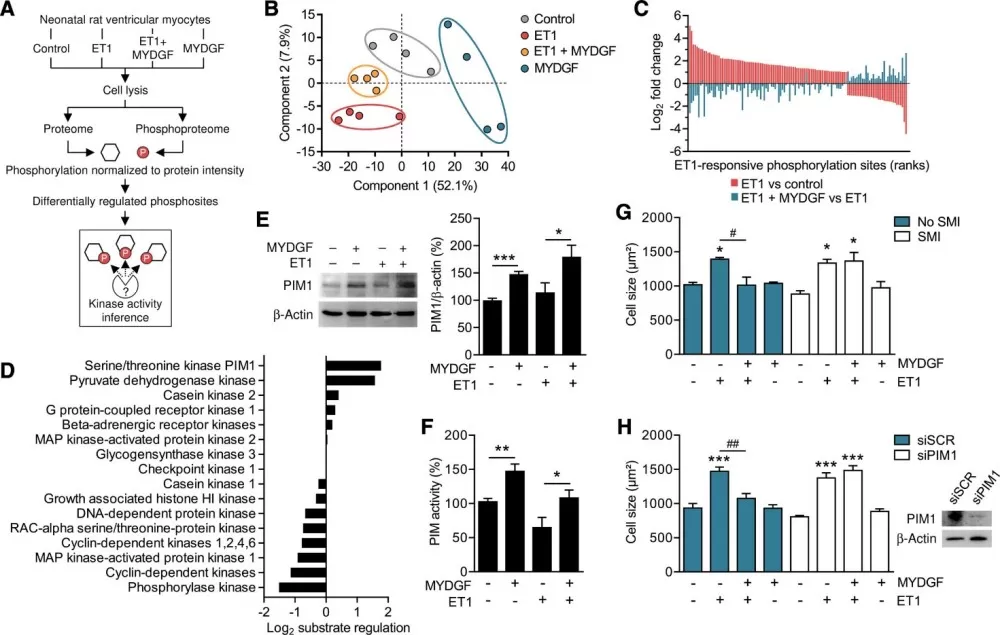
Phosphoproteome analysis identifies PIM1 as a signaling target of MYDGF
MYDGF Therapy Attenuates Heart Failure in Mice
Subcutaneous administration of recombinant MYDGF in TAC mice increased SERCA2a levels, reduced hypertrophy, and improved survival. Proteomic validation in isolated cardiomyocytes confirmed SERCA2a upregulation, while phosphoproteomics linked these effects to sustained PIM1 activation. The Combined proteomic and functional assays provided a holistic view of MYDGF’s therapeutic potential.
This study exemplifies how multi-omics integration accelerates discovery in cardiovascular research. By pairing conventional proteomics (to track MYDGF and SERCA2a) with phosphoproteomics (to map PIM1 signaling), the authors bridged the gap between immune cell biology and cardiomyocyte function. Such approaches are indispensable for identifying druggable targets and understanding post-translational regulatory networks. Notably, the use of LC-MS and phosphopeptide enrichment techniques ensured high-resolution data, while cross-species validation (mice and humans) strengthened translational relevance. Compared to single-omics studies, this dual strategy offers deeper mechanistic insights, though challenges remain in scaling these methods for clinical applications.
Unlock Precision Cardiology with MetwareBio’s Omics Solutions
At Metware Biosciences, we provide cutting-edge proteomics and phosphoproteomics services to accelerate your research. Our high-resolution LC-MS technology, expert bioinformatics, and ISO-certified workflows deliver precise insights into protein expression and post-translational modifications (PTMs). Whether deciphering kinase signaling, mapping phosphorylation events, or identifying disease biomarkers, our comprehensive solutions empower discoveries in cardiovascular, cancer, and metabolic research. From sample preparation to data interpretation, we offer end-to-end support to ensure high-impact results. Contact us today to explore our quantitative protein and modified protein joint analysis report and see how multi-omics can advance your next breakthrough.
Reference
Korf-Klingebiel M, Reboll MR, Polten F, Weber N, Jäckle F, Wu X, Kallikourdis M, Kunderfranco P, Condorelli G, Giannitsis E, Kustikova OS, Schambach A, Pich A, Widder JD, Bauersachs J, van den Heuvel J, Kraft T, Wang Y, Wollert KC. Myeloid-Derived Growth Factor Protects Against Pressure Overload-Induced Heart Failure by Preserving Sarco/Endoplasmic Reticulum Ca2+-ATPase Expression in Cardiomyocytes. Circulation. 2021 Oct 12;144(15):1227-1240. doi: 10.1161/CIRCULATIONAHA.120.053365.


