Unveiling Protein SUMOylation: Key Mechanisms and Biological Significance
Protein SUMOylation, also known as Small Ubiquitin-like Modifier (SUMO) modification, is a crucial post-translational modification that falls under the category of ubiquitin-like modifications. It involves the covalent attachment of SUMO to specific lysine residues on target proteins, leading to changes in the protein's function, stability, localization, or interactions with other molecules.
SUMOylation occurs in various cellular contexts, including responses to environmental changes and stress, regulation of the cell cycle and division, protein localization and transport, protein stability and degradation, protein-protein interactions, and signal transduction. It also plays roles in specific physiological and pathological processes. Understanding the mechanisms and functions of SUMOylation provides valuable insights into cellular activities and the molecular basis of disease development.
Chemical Process of SUMOylation
SUMOylation is catalyzed by a series of enzymes, including E1 SUMO-activating enzymes, E2 SUMO-conjugating enzymes, and in certain cases, E3 SUMO ligases. The process involves several key steps:
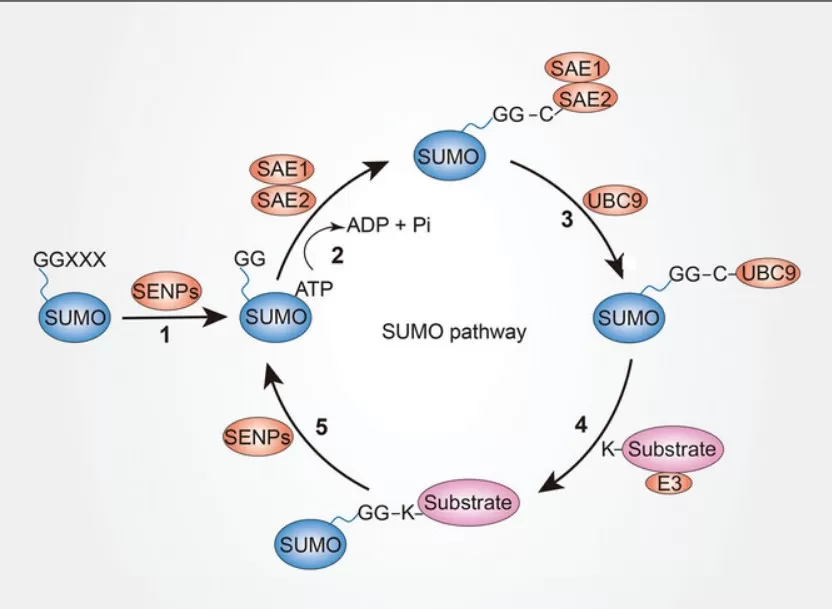
1) Maturation of SUMO precursor: After synthesis, SUMO proteins undergo specific cleavage to generate their mature form.
2) Activation: The mature SUMO is activated by E1 enzymes (activating enzymes). E1 catalyzes the formation of a thioester bond between the C-terminus of SUMO and the cysteine residue on E1, thereby activating SUMO.
3) Conjugation: The activated SUMO is transferred to the target protein via E2 enzymes (conjugating enzymes, such as Ubc9). The E2 enzyme positions SUMO near the lysine residue of the target protein.
4) Ligation (optional): In some cases, E3 ligases are involved to enhance the efficiency of SUMO attachment to the target protein. E3 enzymes specifically recognize substrate proteins and facilitate the formation of an isopeptide bond between SUMO and the target protein.
Roles and Regulatory Mechanisms of SUMOylation
SUMOylation plays a broad role in cellular biology, with its regulatory mechanisms spanning multiple levels:
Regulation of Protein Function
SUMOylation can alter protein functionality in various ways, such as modifying interactions between proteins or with DNA, and sometimes by blocking ubiquitination sites. It also affects protein subcellular localization, as many SUMOylated proteins display SUMO-dependent distribution. For example, SUMOylation regulates the function of poly(A) polymerase (PAP), enhancing its nuclear localization, stability, and activity.
SUMOylation is also critical in disease states. In Parkinson's disease (PD), the accumulation and aggregation of α-synuclein are associated with SUMOylation. This modification elevates α-synuclein levels through two mechanisms: PIAS2-mediated accumulation and aggregation, and inhibition of SIAH- and Nedd4-mediated ubiquitination, further promoting aggregation. SUMOylation is also involved in cardiovascular diseases, neurodegenerative disorders, and other pathological conditions.
Involvement in Subcellular Localization
SUMOylation plays a key biological role by influencing protein localization within the cell, such as nuclear-cytoplasmic shuttling and membrane binding. These effects extend beyond basic cellular processes, like transcription regulation and protein stability, to more complex physiological and pathological processes, including cell migration and disease progression.
Regulation of Gene Expression
SUMOylation regulates gene expression through several mechanisms, including directly modifying specific transcription factors or co-regulatory proteins, impacting their stability and degradation, and influencing their interactions with DNA or other proteins.
Response to Environmental Stress
Under environmental stress conditions such as heat shock or oxidative stress, SUMOylation helps cells cope by maintaining homeostasis and enhancing stress response pathways.
Common Methods for Analyzing SUMOylation
1) Western Blot Analysis: This method detects the presence of SUMOylated proteins in a sample using specific antibodies. Typically, antibodies targeting SUMO proteins or potential substrate proteins are used in immunoblotting after separating proteins via electrophoresis.
2) Immunoprecipitation (IP): Immunoprecipitation involves using specific antibodies, such as anti-SUMO or anti-target protein antibodies, to bind protein-protein complexes in cell lysates. Through subsequent washing and elution steps, these complexes can be purified. This technique helps identify proteins that interact with SUMOylated proteins and confirms their SUMOylation status.
3) Mass Spectrometry (MS): Mass spectrometry, especially liquid chromatography-tandem mass spectrometry (LC-MS/MS), is a powerful tool for identifying and quantifying protein modifications. In SUMOylation analysis, MS can accurately determine the modification sites (specific lysine residues) and the abundance of SUMOylation. Protein samples are typically digested into peptides before being analyzed by MS.
4) In Vivo and In Vitro SUMOylation Assays: In in vivo assays, genetic manipulations like overexpression or knockout of SUMO-related enzymes are used to observe changes in SUMOylation within cells. In vitro assays involve using purified E1, E2, and (optionally) E3 enzymes, along with target proteins, to reconstitute the SUMOylation reaction outside the cell, allowing for the study of its molecular mechanisms and specificity.
5) Bioinformatics Analysis: Bioinformatics tools are employed to predict potential SUMOylation sites. These tools are typically based on the sequence characteristics of known SUMOylated proteins, such as conserved motifs around SUMOylation sites. While these predictions require experimental validation, they provide useful guidance for selecting candidate proteins for further study.
Reference
1. Zhou H,Deng N,Li Y, et al. Distinctive tumorigenic significance and innovative oncology targets of SUMOylation. Theranostics. 2024;14 (8):3127-3149. doi:10.7150/thno.97162
2. Vethantham V,Rao N,Manley JL. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 2008;22 (4):499-511. doi:10.1101/gad.1628208
3. Prasad S,Pal PK. SUMOylation: One small modification for proteins, multiple giant problems for mankind. Mov Disord. 2018;33 (3):403. doi:10.1002/mds.27293
4. Du C,Chen X,Su Q, et al. The Function of SUMOylation and Its Critical Roles in Cardiovascular Diseases and Potential Clinical Implications. Int J Mol Sci. 2021;22 (19):doi:10.3390/ijms221910618
5. Erazo T,Espinosa-Gil S,Diéguez-Martínez N, et al. SUMOylation Is Required for ERK5 Nuclear Translocation and ERK5-Mediated Cancer Cell Proliferation. Int J Mol Sci. 2020;21 (6):null. doi:10.3390/ijms21062203
6. Gong L,Ji WK,Hu XH, et al. Sumoylation differentially regulates Sp1 to control cell differentiation. Proc Natl Acad Sci U S A. 2014;111 (15):5574-9. doi:10.1073/pnas.1315034111
7. Li W,Liu W,Xu Z, et al. Heat-induced SUMOylation differentially affects bacterial effectors in plant cells. Plant Cell. 2024;36 (6):2103-2116. doi:10.1093/plcell/koae049
8. Fan, Y., Li, X., Zhang, L., Zong, Z., Wang, F., Huang, J., Zeng, L., Zhang, C., Yan, H., Zhang, L., & Zhou, F. (2022). SUMOylation in Viral Replication and Antiviral Defense. Advanced science (Weinheim, Baden-Wurttemberg, Germany), 9(7), e2104126. https://doi.org/10.1002/advs.202104126
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


