The Fine Art of Protein Methylation: Mechanisms, Impacts, and Analytical Techniques
In the microscopic world of cells, proteins play an extremely important role. They are not only key components of cell structures but also participate in nearly all biochemical reactions. The function and activity of proteins can be regulated in various ways, one of which is protein methylation. Protein methylation is a sophisticated regulatory mechanism within cells, profoundly impacting cellular functions and interactions by modifying protein activity. It plays a critical role in cellular signal transduction, gene expression regulation, and the progression of diseases. Understanding the mechanisms and functions of protein methylation not only enhances our comprehension of fundamental principles in life sciences but also opens avenues for developing new drugs and therapeutic strategies. Today, we will explore the mechanisms, types, functions and analytical techniques of protein methylation.
What is Protein Methylation?
Protein methylation is a common type of post-translational modification (PTM) where a methyl group (-CH₃) is transferred from S-adenosyl-L-methionine (SAM) to specific amino acid residues by methyltransferases. This modification can occur on both histone and non-histone proteins, typically at arginine (Arg) and lysine (Lys) residues. Protein methylation not only influences protein stability, subcellular localization, and enzymatic activity, but it can also alter interactions between proteins as well as their binding capabilities with DNA or RNA.
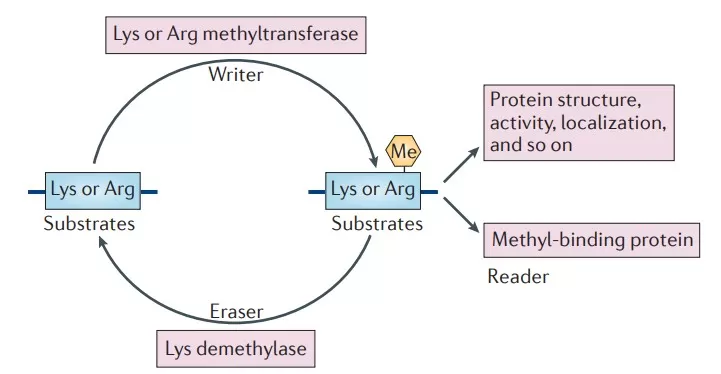
Mechanism of Protein Methylation
The occurrence of protein methylation is influenced by various factors, including the availability of enzymes and the supply of methyl donors. This process is catalyzed by a specialized class of enzymes known as methyltransferases (MTases). Methyltransferases play a crucial role in this process as they can specifically recognize and transfer methyl groups to target amino acid residues. For example, METTL9 is a histidine N1 methyltransferase that can identify substrates and catalyze the N1 methylation of histidine. Additionally, demethylases, such as lysine-specific demethylases and members of the Jumonji domain-containing demethylase family, are responsible for removing methylation modifications.
Types of Protein Methylation
1. Monomethylation: A single methyl group is added to an amino acid residue.
2. Dimethylation: Two methyl groups are added to the same amino acid residue.
3. Trimethylation: Three methyl groups are added to the same amino acid residue, representing the highest level of methylation.
Protein methylation primarily occurs on lysine and arginine residues, and it can manifest as monomethylation, dimethylation, or trimethylation. Common methylation sites on histones include H3-K4, H3-K9, and H3-K27; these methylation states significantly influence chromatin structure and gene expression. Methylation on non-histone proteins is also critical; for instance, METTL9 maintains zinc ion homeostasis by methylating the zinc transporter SLC39A7, facilitating tumor cell growth.
Effects of Protein Methylation
- Regulation of Gene Expression: Methylation on histones can alter chromatin structure and DNA accessibility, thus regulating gene transcription. For example, trimethylation of lysine 4 on histone H3 (H3K4me3) is typically associated with gene activation, while trimethylation of lysine 9 (H3K9me3) correlates with gene silencing.
- Signal Transduction: Protein methylation can modulate key proteins within signaling pathways, affecting how cells respond to external signals. The methylation status of certain transcription factors and signaling molecules can determine their activity and stability.
- Protein Stability: Methylation can influence the rate of protein degradation. Some proteins become more stable following methylation, while others may be more susceptible to degradation due to this modification.
- Protein-Protein Interactions: Methylation can alter the surface charge and conformation of proteins, impacting their ability to bind with other proteins.
Role of Protein Methylation in Diseases
- Dysregulation of protein methylation is linked to the development of various diseases, including cancer, neurodegenerative disorders, and cardiovascular diseases.
- Cancer: Changes in methylation patterns can lead to the silencing of tumor suppressor genes or the activation of oncogenes in certain cancers.
- Neurodegenerative Diseases: Abnormal methylation of neurotransmitter receptors and signaling molecules may be associated with neurodegenerative diseases and mental disorders.
- Cardiovascular Diseases: Alterations in protein methylation in some cardiovascular diseases can affect vascular contraction and relaxation, thereby influencing blood pressure and flow.
Techniques for Studying Methylation
1. Mass Spectrometry (MS)
Mass spectrometry (MS) is a highly effective analytical technique employed to identify and quantify methylated proteins and their modifications at the molecular level. This method can precisely determine which specific residues, such as lysine (Lys) and arginine (Arg), are methylated, as well as quantify the levels of methylation. This capability offers valuable insights into the biological significance of changes in methylation patterns. The strengths of MS analysis include its high sensitivity and specificity, the ability to analyze complex mixtures, and the provision of detailed structural information regarding methylation modifications. The key steps involove:
1) Sample Preparation: Proteins are often digested into peptides using proteolytic enzymes (e.g., trypsin).
2) Liquid Chromatography (LC): Peptides are separated using liquid chromatography before entering the mass spectrometer.
3) Ionization: Techniques such as Electrospray Ionization (ESI) or Matrix-Assisted Laser Desorption/Ionization (MALDI) are used to ionize the peptides.
4) Mass Analysis: The mass-to-charge ratio (m/z) of the ionized peptides is measured. Different methylation states will produce distinct mass shifts, which can be analyzed.
5) Data Interpretation: Software tools are used to analyze the mass spectra and identify specific methylated sites based on their mass differences.
2. Western Blotting
Western blotting is a widely used method for detecting specific proteins in a sample, and it can be modified to identify methylated proteins effectively. This technique is valuable for verifying the presence of specific methylated proteins and quantifying their levels, enabling comparative studies of methylation status between different samples or conditions. One of the key strengths of Western blotting is its relative simplicity and cost-effectiveness, along with the ability to detect multiple proteins in a single experiment, making it a versatile tool in protein methylation research. The key steps involove:
1) Sample Preparation: Proteins are extracted and quantified from cells or tissues.
2) Gel Electrophoresis: Proteins are separated based on their size using SDS-PAGE.
3) Transfer: Separated proteins are transferred to a membrane (e.g., nitrocellulose or PVDF).
4) Blocking: The membrane is blocked to prevent non-specific binding of antibodies.
5) Antibody Incubation: The membrane is incubated with specific antibodies against methylated residues (e.g., anti-H3K4me3).
6) Detection: The bound antibodies are visualized using chemiluminescent or colorimetric substrates.
3. Immunoprecipitation (IP)
Immunoprecipitation is a technique utilized to isolate specific proteins or protein complexes from a mixture, facilitating the study of methylated proteins. This method is particularly valuable for identifying interacting partners, revealing other proteins that interact with the methylated protein of interest, and assessing the functional implications of methylation on protein interactions. One of the key strengths of immunoprecipitation is its high specificity for the protein of interest, allowing for the enrichment of low-abundance methylated proteins, which can be crucial for understanding their biological roles. The key steps involove:
1) Cell Lysis: Cells are lysed to release proteins.
2) Antibody Binding: Specific antibodies against the target protein or methylation mark are added to the lysate, allowing them to bind the desired protein.
3) Capture: Protein-A/G beads are used to capture the antibody-protein complexes.
4) Washing: Unbound proteins are washed away.
5) Analysis: The captured proteins can be analyzed via Western blotting or mass spectrometry.
Reference
1. Biggar, K. K., & Li, S. S. (2015). Non-histone protein methylation as a regulator of cellular signalling and function. Nature reviews. Molecular cell biology, 16(1), 5–17. https://doi.org/10.1038/nrm3915
2. Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010 Jul;10(7):457-69. doi: 10.1038/nrc2876.
3. Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012 Apr 3;13(5):343-57. doi: 10.1038/nrg3173.
4. Lim Y, Lee JY, Ha SJ, Yu S, Shin JK, Kim HC. Proteome-wide identification of arginine methylation in colorectal cancer tissues from patients. Proteome Sci. 2020 May 19;18:6. doi: 10.1186/s12953-020-00162-8.
5. Sun Z, Liu L, Chen J. Targeting non-histone methylation in gastrointestinal cancers: From biology to clinic. Eur J Med Chem. 2024 Aug 26;278:116802. doi: 10.1016/ j.ejmech. 2024. 116802.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


