Enhancing Proteomics Analysis: A Comprehensive Guide to Protein Digestion and Desalting
In previous blogs, we discussed how to collect proteomics samples and extract proteins from various sample types. Once the proteins are successfully extracted, they must undergo two critical steps: enzymatic digestion and desalting. Enzymatic digestion breaks down complex proteins into smaller peptides, making them more suitable for mass spectrometry (MS) analysis, while desalting removes interfering substances like salts and small molecules that can affect the accuracy and sensitivity of MS detection. These steps are crucial in ensuring high-quality, reproducible data in proteomics studies. In this blog, we will explore the different methods used for digestion and desalting, providing insights into how to optimize these processes for various types of proteomics samples. Whether working with large clinical samples like blood serum or more challenging fluids like cerebrospinal or urine, an understanding of these techniques is essential for obtaining the most accurate proteomic profiles.
Breaking Down Proteins: Key Enzymatic Techniques for Proteomics
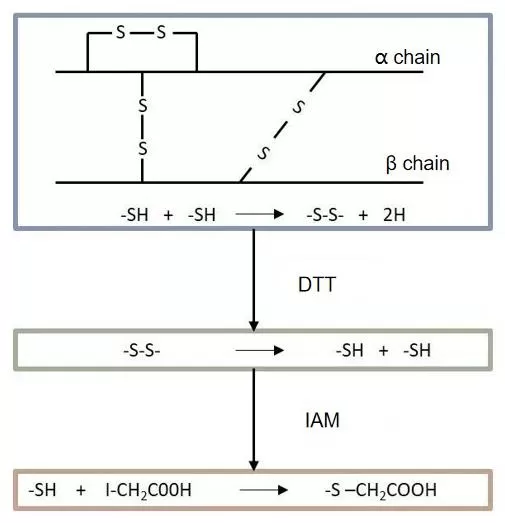
Protein digestion aims to produce peptides suitable for mass spectrometry (MS) detection, making it a critical step in sample preparation. Several factors, such as the digestion system, enzyme type, enzyme amount, and digestion time, directly affect the digestion efficiency and protein identification results. Reduction and alkylation are key components of this process. Dithiothreitol (DTT) is commonly used to reduce protein disulfide bonds, preventing cysteines from forming new disulfide bridges, while iodoacetamide (IAA) alkylates the reduced thiol groups, keeping them from re-oxidizing and ensuring full protein denaturation to improve digestion efficiency (see Fig. 1).
Trypsin, the most widely used protease, is highly specific and cleaves peptide bonds at the C-terminal of lysine (K) and arginine (R), unless proline (P) is adjacent to the cleavage site. It operates most efficiently at pH 7–9 and becomes inactive below pH 4. Trypsin digestion produces peptides that are typically 2+ charged and of ideal length for MS analysis. However, for some proteins with few cleavage sites, trypsin alone may produce peptides that are too long for effective MS detection. In such cases, combining trypsin with other enzymes, like Lys-C, can shorten the peptides and increase the identification rate in proteomic analysis.
Two digestion methods can be applied based on total protein amount: for samples with over 200 µg, precipitation digestion is recommended, while for amounts under 200 µg, in-solution digestion is preferred.
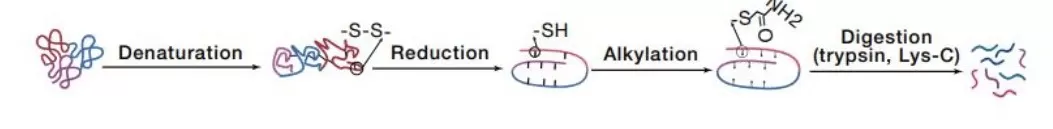
The following outlines the general steps of in-solution digestion (see Fig. 2):
1. Take 30 µg of protein and adjust the volume to 100 µL with 8M urea.
2. Add DTT to a final concentration of 5 mM, incubate at 37°C for 45 minutes for reduction.
3. Add IAA to a final concentration of 11 mM, incubate in the dark for 15 minutes, then stop the reaction with light exposure. The reduction and alkylation steps are now complete.
4. Add 400 µL of TEAB to bring the volume to 500 µL.
5. Add trypsin at a 1:75 ratio and incubate at 37°C overnight for digestion.
6. To stop the digestion, acidify the solution with 20% FA to a pH of about 3.
7. Finally, perform desalting using a C18 column.
Cleaning Peptides: Essential Desalting Techniques for Accurate MS Detection
Before mass spectrometry, desalting of digested peptides is crucial to ensure accurate peptide identification and prevent damage to the instrument. C18 desalting is commonly used in this process. After desalting, peptides are quantified using BCA, and 1-2 µg of the peptide sample is lyophilized, reconstituted in loading buffer to 100 ng/µL, and prepared for MS analysis.
C18 Zip-Tip Desalting Procedure:
1. Activate the C18 Zip-Tip with methanol.
2. Equilibrate twice with 0.1% TFA solution.
3. Load the digested peptide sample onto the C18 Zip-Tip and repeat three times for thorough adsorption.
4. Wash the tip twice with 0.1% TFA to remove impurities.
5. Elute the peptides in two steps:
6. First, with 20 µL of 50% ACN in 0.1% formic acid, repeat once.
7. Then, with 20 µL of 80% ACN in 0.1% formic acid. The collected eluent contains desalted peptides ready for mass spectrometry analysis.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


