Protein Characterization: Unveiling the Mysteries of Life Molecules
Introduction to Protein Characterization
Protein characterization is a cornerstone of biomedical research and biotechnology, enabling scientists to decipher the structure, function, and interactions of proteins, thereby unlocking the secrets of life. Understanding protein structure and function is pivotal for advancing drug discovery, designing biomaterials, and elucidating disease mechanisms. Protein characterization refers to the integration of biochemical, physicochemical, and molecular biological methods to determine and describe a protein’s structure, properties, and functions. This process reveals functional mechanisms, identifies disease-associated mutations, guides protein engineering, and optimizes industrial enzymatic processes.
As a multidisciplinary field intersecting biochemistry, biophysics, and computational biology, protein characterization encompasses analyses at multiple levels: molecular weight, structural hierarchy (primary to quaternary), functional activity, purity, post-translational modifications (PTMs), and dynamic regulatory changes.
Molecular Weight Determination
Accurate molecular weight determination is foundational for protein characterization. Mass spectrometry (MS) is the gold standard, measuring the mass-to-charge ratio (m/z) of ionized proteins to provide precise molecular weight data. Complementary techniques like gel permeation chromatography (GPC) and ultrafiltration offer indirect estimates based on hydrodynamic volume or size exclusion. These methods are indispensable in both basic research and biopharmaceutical development, aiding in the validation of protein composition and structural integrity. Recent advancements in high-resolution MS, such as Orbitrap and time-of-flight (TOF) analyzers, now enable sub-Dalton accuracy, even for large protein complexes.
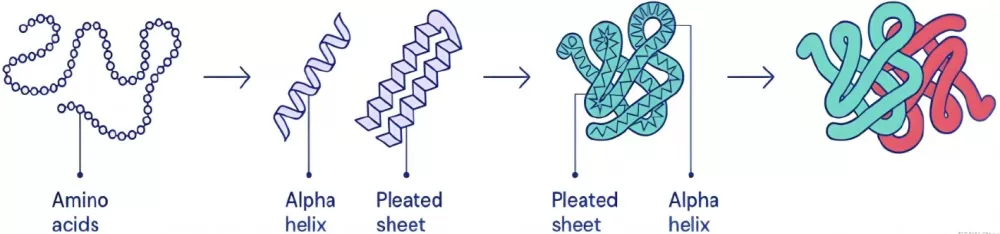
Protein Structural Characterization
Structural Characterization: Decoding Protein Architecture
1. Primary Structure Analysis
The primary structure, defined by the linear sequence of amino acids, underpins a protein’s identity and evolutionary relationships. Techniques like Edman degradation (sequential N-terminal amino acid cleavage) and tandem MS (peptide fragmentation and sequencing) are widely used. For example, MS-based ‘de novo sequencing’ bypasses the need for genomic data, making it invaluable for characterizing novel proteins.
2. Secondary and Tertiary Structure Analysis
Secondary structures (α-helices, β-sheets) and tertiary structures (3D folding) dictate protein stability and functional motifs. X-ray crystallography resolves atomic-level details using protein crystal diffraction, while nuclear magnetic resonance (NMR) spectroscopy captures dynamic structures in solution. Cryogenic electron microscopy (Cryo-EM) excels in resolving large complexes (>150 kDa) at near-atomic resolution, revolutionizing structural biology.
3. Quaternary Structure Analysis
Quaternary structures involve subunit assembly in multimeric proteins. Cross-linking mass spectrometry (XL-MS) identifies spatial proximities between subunits, whereas fluorescence resonance energy transfer (FRET) monitors real-time interactions. These methods elucidate functional oligomerization, as seen in hemoglobin’s oxygen-binding cooperativity.
Functional Characterization: Understanding Protein Activity
1. Enzymatic Activity Studies
Enzymes catalyze biochemical reactions with remarkable specificity. Kinetic parameters like the Michaelis constant (Km) and maximum velocity (Vmax) are determined via spectrophotometric assays. For instance, inhibitors targeting HIV protease’s active site have been optimized using enzyme kinetics, highlighting its therapeutic relevance.
2. Binding Affinity and Specificity
Protein-ligand interactions are quantified using surface plasmon resonance (SPR) and biolayer interferometry (BLI), which measure real-time binding kinetics (e.g., dissociation constant, *Kd*). These techniques are critical in drug discovery, such as characterizing monoclonal antibody-antigen interactions.
3. Functional Screening
High-throughput mutagenesis and CRISPR-Cas9 editing identify critical residues or domains. For example, alanine scanning of tumor suppressor p53 revealed residues essential for DNA binding and tumor suppression.
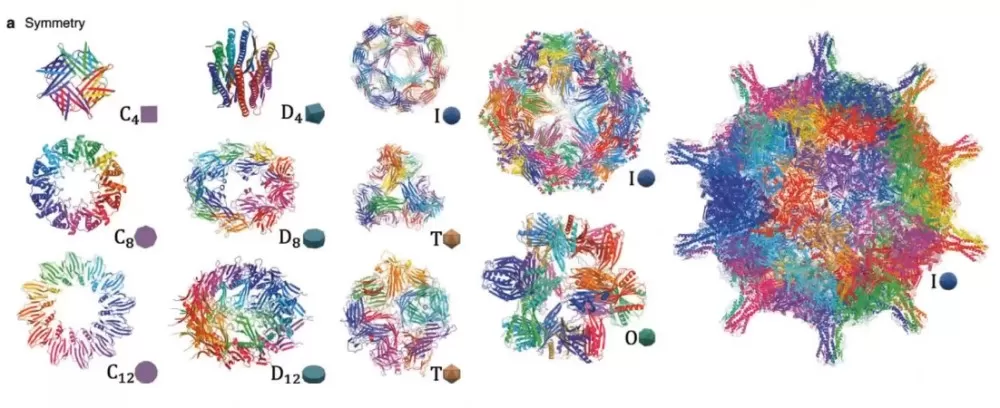
protein structure generation
Purity Analysis
Protein purity is paramount for reproducible research. SDS-PAGE separates proteins by molecular weight, while reverse-phase HPLC and capillary electrophoresis detect impurities with high sensitivity. Two-dimensional gel electrophoresis (2D-GE) combines isoelectric focusing (IEF) and SDS-PAGE to resolve isoforms and PTM variants. Modern MS-based approaches, like intact protein analysis, now detect contaminants at ppm levels, ensuring compliance with biopharmaceutical quality standards.
Dynamic Changes and Regulatory Characterization
Proteins undergo dynamic regulation through post-translational modifications (PTMs) and participate in intricate interaction networks, both of which are critical for their functional diversity and cellular roles. PTMs, such as phosphorylation, ubiquitination, and glycosylation, modulate protein activity, localization, and stability. For example, phosphoproteomics using titanium dioxide enrichment and mass spectrometry (MS) has identified key kinase signaling hubs in cancer, while glycoproteomics has mapped glycosylation sites essential for antibody effector functions. Beyond PTMs, proteins operate within complex interaction networks that govern cellular processes. Techniques like yeast two-hybrid (Y2H) screens and co-immunoprecipitation (Co-IP) coupled with MS have been instrumental in uncovering these networks. A notable example is the SARS-CoV-2 interactome, which revealed host proteins co-opted by the virus for replication, providing critical insights for antiviral drug design. Together, the study of PTMs and interaction networks offers a comprehensive understanding of protein regulation and its implications in health and disease.
Applications and Future Perspectives in Protein Science
Protein characterization drives innovations across diverse fields, including drug development, precision medicine, and industrial biotechnology. In drug development, engineered antibodies such as checkpoint inhibitors rely on detailed epitope mapping and affinity maturation to enhance therapeutic efficacy. In precision medicine, mutational profiling of oncoproteins like EGFR and BRAF informs the design of targeted therapies, improving treatment outcomes for cancer patients. In industrial biotechnology, enzyme engineering optimizes biocatalysts for applications such as biofuel production, enhancing efficiency and sustainability. Emerging technologies like single-molecule imaging and AI-driven structure prediction (e.g., AlphaFold) are revolutionizing the field by accelerating characterization workflows and bridging the gaps between protein sequence, structure, and function. These advancements promise to unlock new possibilities in understanding protein biology and developing innovative solutions for global challenges.
Protein characterization is an ever-evolving discipline central to life sciences. By integrating cutting-edge analytical tools and computational models, researchers continue to decode the complexities of proteins, paving the way for transformative discoveries in health, industry, and beyond.
Read more
- What is Phospholipid? Structure, Functions, and Applications
- Unveiling Ornithine: Beyond the Urea Cycle, A Multifaceted Player in Health
- Top-Down vs. Bottom-Up Proteomics: Unraveling the Secrets of Protein Analysis
- Comprehensive Guide to Basic Bioinformatics Analysis in Proteomics
- Strategies for Selecting Key Interacting Proteins in IP-MS Studies
- Peptide and Protein De Novo Sequencing by Mass Spectrometry
- How to understand the WGCNA analysis in publications? (1/2)
- Understanding WGCNA Analysis in Publications
- Harnessing the Power of WGCNA Analysis in Multi-Omics Data


