The Hidden Regulators: An In-Depth Look at Post-Translational Modifications in Proteins
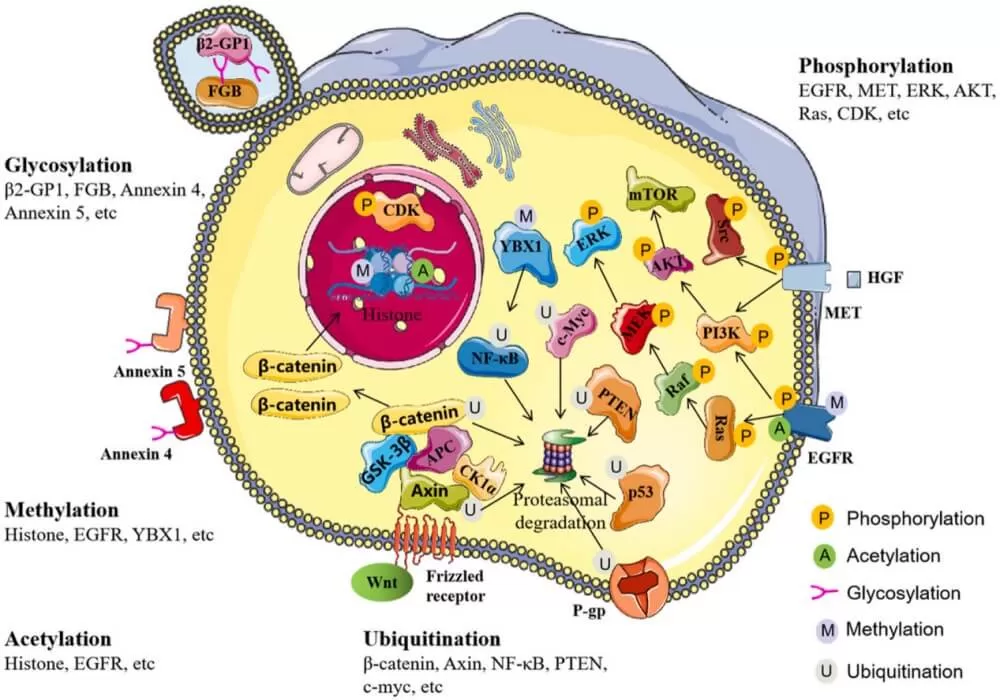
Post-translational modification (PTM) refers to the process by which chemical groups are added to or removed from specific amino acid residues of proteins after synthesis through enzymatic reactions. Common modifications include phosphorylation, methylation, and acetylation. These modifications regulate the three-dimensional structure, function, interactions, stability, and intracellular localization of most eukaryotic proteins, acting as rapid and reversible switches to efficiently modulate protein activity. Extensive research has shown that aberrant regulation of PTMs is closely associated with various human diseases.
Post-translational modifications are typically catalyzed by enzymes, but some studies suggest that certain PTMs can occur spontaneously without enzymatic involvement. These non-enzymatic PTMs can be categorized into two main types based on their stability and physiological lifespan: irreversible and reversible non-enzymatic PTMs. Irreversible non-enzymatic PTMs, such as modifications induced by oxidative stress and metabolic stress, cannot be removed through spontaneous dissociation or enzymatic mechanisms. In contrast, reversible non-enzymatic PTMs can be reversed under physiological conditions, either through enzymatic catalysis or due to inherent chemical instability.
These processes are involved in various metabolic and signaling pathways The traditional PTMs such as phosphorylation, acetylation, ubiquitination, and methylation have been extensively studied, newly discovered modifications like hydroxybutyrylation, lactylation, and succinylation offer fresh insights into the regulatory mechanisms of proteins. These novel modifications also open new avenues for exploring their potential applications in biological systems.
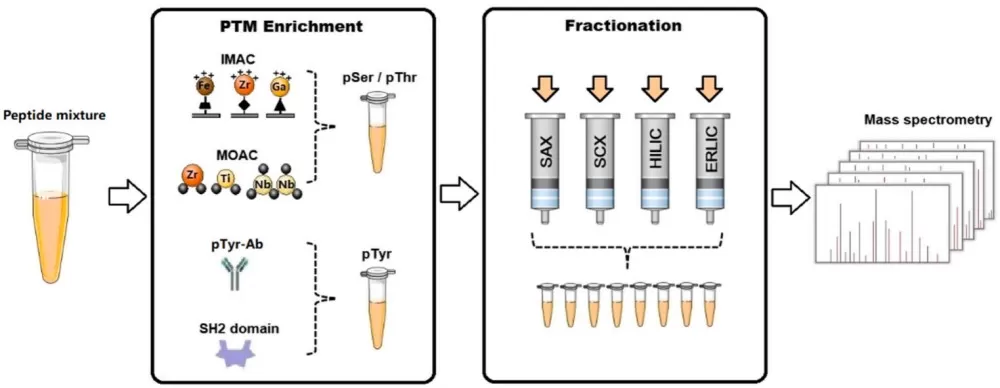
Since PTMs typically occur at low abundance within the proteome, detecting or quantifying them without prior enrichment can be challenging. To address this, scientists have developed a range of enrichment strategies tailored to specific PTMs. Once enriched, these modified proteins are analyzed using proteomics techniques, with mass spectrometry (MS) being the primary tool for characterizing the specific mass shifts of precursor ions and the mass signatures of fragment ions. In this blog, we will provide a detailed overview of various types of post-translational modifications, along with their commonly used enrichment and detection methods.
Phosphorylation
Phosphorylation is a widely prevalent post-translational modification found across various organisms, frequently occurring on proteins within both the nucleus and cytoplasm. This process is catalyzed by kinases, which transfer a phosphate group from ATP to specific amino acid residues, such as serine (Ser), threonine (Thr), tyrosine (Tyr), and histidine (His). Phosphorylation regulates protein function either through allosteric effects or by mediating interactions with other domains. Phosphorylation and dephosphorylation are reversible enzymatic reactions, catalyzed by kinases and phosphatases, respectively.
Given that phosphorylated proteins are typically low in abundance, specific enrichment methods are required to enhance detection sensitivity prior to mass spectrometry (MS) analysis. Common enrichment techniques include metal oxide affinity chromatography (MOAC), immobilized metal ion affinity chromatography (IMAC), immunoaffinity enrichment, and domain-specific enrichment. To further reduce sample complexity, various separation strategies are employed, such as strong cation exchange chromatography (SCX), strong anion exchange chromatography (SAX), hydrophilic interaction liquid chromatography (HILIC), and electrostatic repulsion hydrophilic interaction chromatography (ERLIC). The enriched phosphopeptides can then be identified and quantified using MS in combination with stable isotope labeling techniques, such as SILAC, iTRAQ, and TMT.
Acetylation
Acetylation is a reversible protein modification process, catalyzed by lysine acetyltransferases (KATs), which transfer an acetyl group from acetyl-CoA to the lysine residues of target proteins. This process can be reversed by deacetylases (KDACs), which play crucial roles in chromatin remodeling, gene expression regulation, and the modulation of protein function. In addition to histones, many non-histone proteins are responsive to acetylation and other post-translational modifications (PTMs). Aberrant acetylation has been observed in various diseases, particularly in cancer, where it is associated with the enhanced glycolytic activity of tumor cells.
Mass spectrometry (MS), especially the top-down proteomics approach, has become a primary tool for identifying and quantifying protein acetylation. Since acetylated proteins are typically low in abundance, enriching and isolating acetylated peptides is a critical step to enhance detection sensitivity. Immunoaffinity purification using antibodies against acetylated lysine is a highly effective method for enriching acetylated peptides, thereby increasing the depth of MS analysis. Strong cation exchange chromatography (SCX) is a commonly used separation technique, often combined with immunoaffinity purification to improve the accuracy of acetylation site identification and quantification.
Glycosylation
Glycosylation is a common form of post-translational modification, primarily categorized into N-glycosylation and O-glycosylation. N-glycosylation occurs on asparagine residues, while O-glycosylation takes place on serine or threonine residues. This multi-step enzymatic process, mainly occurring in the endoplasmic reticulum and Golgi apparatus, is crucial for signal transduction, immune responses, and various biological processes related to cancer. Abnormal glycosylation has been linked to numerous diseases, including cancer and COVID-19, where it holds significant value as a biomarker for disease diagnosis and treatment. Systematic analysis of glycoproteins using mass spectrometry (MS) can shed light on the role of glycosylation in disease progression, such as colorectal cancer, and help in identifying novel biomarkers.
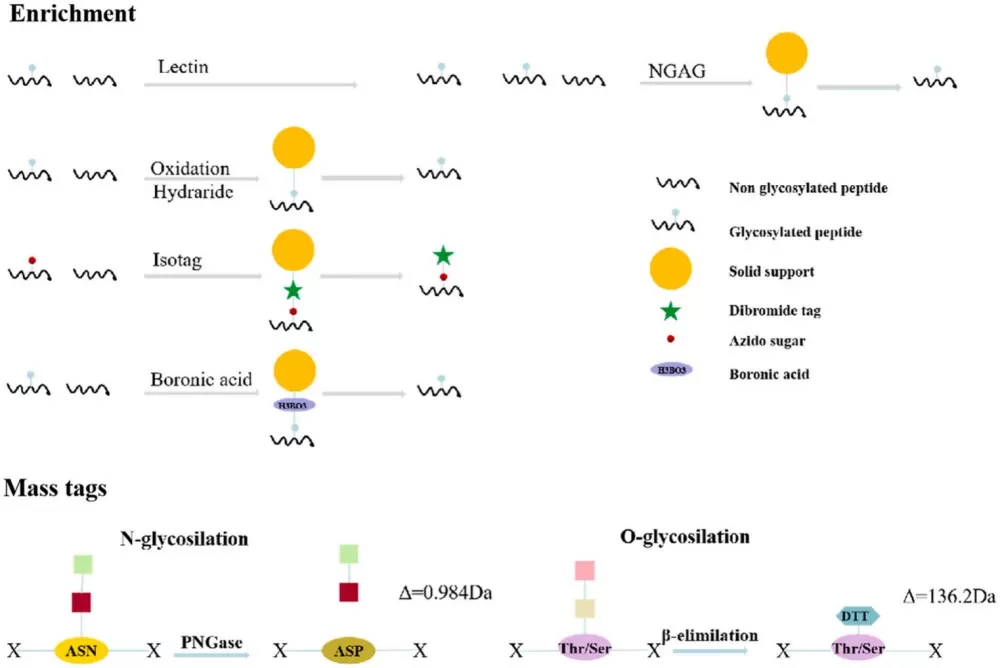
In global glycoprotein analysis, the enrichment of glycopeptides is a critical step. Traditional enrichment methods, such as lectin affinity chromatography and chemical acetylation, simplify the analysis but may result in the loss of important glycan structural information. With technological advancements, MS-based glycoproteomics enrichment strategies have evolved, focusing on glycan types and glycosylation sites. Emerging enrichment techniques include Isotope-Targeted Glycoproteomics (IsoTaG), solid-phase extraction of N-glycans and glycopeptides (NGAG), and boronic acid-based chemical enrichment methods. N-glycosylated peptides are typically analyzed through PNGase F digestion or chemical deglycosylation, while O-glycosylated peptides are produced via β-elimination.
Ubiquitination
Ubiquitin (Ub) is a small protein composed of 76 amino acids that covalently attaches to target proteins through a series of enzymatic reactions involving ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3). This conserved post-translational modification process is known as ubiquitination. Ubiquitin can bind to substrates in various ways, including the attachment of a single ubiquitin molecule to a single amino acid residue (monoubiquitination), the attachment of multiple ubiquitin molecules to different residues (multi-monoubiquitination), or the formation of ubiquitin chains (polyubiquitination). Polyubiquitin chains are created via isopeptide bonds between ubiquitin and the N-terminal methionine (M1) or one of seven internal lysine residues (K6, K11, K27, K29, K33, K48, K63) of the substrate, leading to different structural configurations. Both monoubiquitination and polyubiquitination have significant impacts on protein function under physiological and pathological conditions.
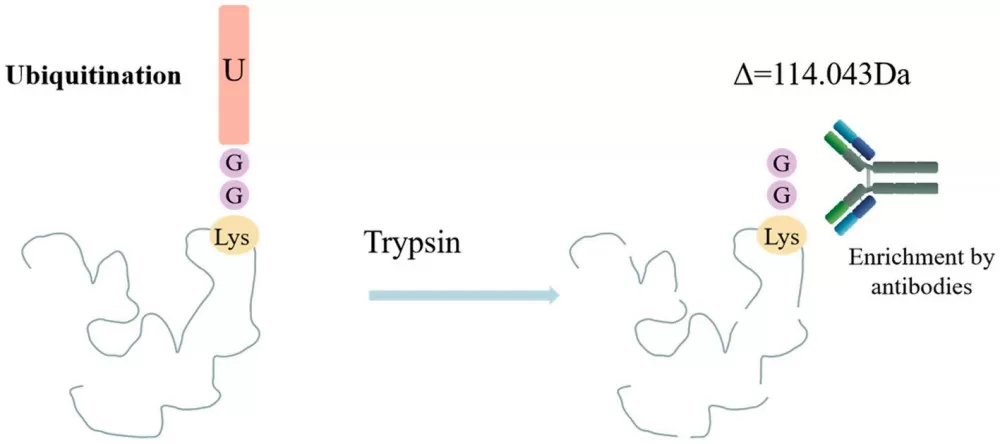
Mass spectrometry (MS) is an effective tool for analyzing ubiquitination, enabling the large-scale identification of ubiquitination sites. During MS analysis, ubiquitinated lysine residues retain a diglycine (K-ε-GG) remnant, resulting in a mass shift of 114.043 Da, which serves as a signature for detection. However, challenges in MS analysis include the low abundance of ubiquitinated peptides and the complexity of multiple ubiquitination sites. Immunoaffinity purification, especially with K-ε-GG antibodies, is an effective strategy for enriching ubiquitinated peptides. Another enrichment method involves introducing affinity tags to the N-terminus of ubiquitin molecules, followed by affinity chromatography to isolate ubiquitinated proteins. This approach can identify a large number of ubiquitinated proteins but may not be suitable for tissue samples.
Methylation
Protein methylation typically involves the transfer of a methyl group from S-adenosylmethionine (SAM) to specific amino acid residues, such as arginine and lysine. This process is catalyzed by protein arginine methyltransferases (PRMTs) and protein lysine methyltransferases (PKMTs), respectively. Although protein methylation has not been as extensively studied as phosphorylation, acetylation, or ubiquitination, it has been shown to play crucial roles in RNA metabolism, gene expression regulation, and protein function. Histones, particularly H3 and H4, are major substrates for methylation, where they regulate chromatin structure and gene transcription. Additionally, the methylation of non-histone proteins, such as NF-κB, p53, pCAF, ERα, and various transcription factors, has been shown to influence protein stability, activity, and function—key processes associated with tumorigenesis, immune responses, and inflammation during cancer and infectious diseases.
Comprehensive analysis of protein methylation presents technical challenges, as methylation induces minimal changes in the physical and chemical properties of peptides, making it difficult to enrich and isolate methylated peptides. Immunoaffinity purification, particularly with antibodies specific to methylated amino acids, is the primary strategy for enriching methylated peptides. This method effectively isolates methylated peptides from complex samples, facilitating subsequent mass spectrometry (MS) analysis and identification of methylation sites.
SUMOylation
SUMOylation is a key post-translational modification process that involves the covalent attachment of small ubiquitin-like modifiers (SUMO) to lysine residues on target proteins through a series of enzymatic reactions. SUMO proteins, approximately 11 kDa in size, are structurally similar to ubiquitin. This process requires the participation of three types of enzymes: SUMO-activating enzymes (SUMO E1), SUMO-conjugating enzymes (SUMO E2), and SUMO ligases (SUMO E3), which work in concert to covalently attach SUMO to the lysine residues of target proteins. Additionally, SUMO proteases, such as sentrin-specific protease 1 (SENP1) in humans, are responsible for removing SUMO from the target proteins.
In many types of malignancies, the expression levels of enzymes involved in the SUMOylation pathway are often elevated, which has been associated with tumorigenesis and poor prognosis in patients. SUMOylation has also been linked to chemotherapy and hormone therapy resistance in several studies. In research on pancreatic cancer, a regulatory relationship between the oncogene Myc and the SUMOylation machinery has been uncovered. These findings suggest that SUMOylation may play a critical role in cancer progression and treatment responses.
Palmitoylation
Palmitoylation is a reversible enzymatic lipid modification in which a 16-carbon palmitic acid is attached to cysteine (Cys) residues via a thioester bond. This process is regulated by enzymes, including the DHHC protein family, which acts as palmitoyl transferases (PATs), and enzymes with depalmitoylating activity such as acyl protein thioesterase (APT) and the α/β-hydrolase domain-containing protein 17 (ABHD17) family. Palmitoylation occurs on many proteins that are closely associated with cancer progression, with the small GTPase RAS family serving as a prime example. Studies have shown that palmitoylation at Cys181 of the NRAS protein is essential for its membrane anchoring, while the removal of this modification can block NRAS downstream signaling, thereby inhibiting the progression of certain types of leukemia.
Succinylation
Succinylation is a durable post-translational modification that occurs when the intermediate metabolite of the Krebs cycle, succinate, reacts with the thiol group of cysteine residues in proteins to form S-(2-succinyl) cysteine (2SC). This modification primarily takes place in the mitochondria of both prokaryotic and eukaryotic organisms, playing a crucial role in mitochondrial energy metabolism. Dysregulated succinylation has been linked to the development of various diseases, including cancer, cardiovascular diseases, liver diseases, and neurological disorders.
Research shows that 2SC serves as a biomarker reflecting mitochondrial stress in conditions such as obesity and diabetes. Furthermore, in patients with hereditary leiomyomatosis and renal cell carcinoma (HLRCC), where fumarate hydratase (FH) is deficient, elevated levels of 2SC are observed. There is also evidence that succinate may be transported from the mitochondria to other parts of the cell or even extracellularly, potentially leading to 2SC formation and damage to proteins outside of the mitochondria. These damaged proteins form what is referred to as the succinylated proteome, which may result in the loss of their original function and structural activity, ultimately leading to cell death. Increasing evidence indicates that protein succinylation is elevated in conditions such as diabetes, obesity, and certain cancers. Thus, 2SC, as a biomarker of mitochondrial stress, may be closely associated with cellular dysfunction and even apoptosis in cancer and diabetes.
Propionylation
Propionylation is an emerging acylation modification of proteins, wherein a propionyl group is covalently attached to lysine residues on target proteins, resulting in propionylated lysine (Kpr). This modification was initially discovered on histones but has since been observed in various non-histone proteins as well. Propionyl-CoA serves as the donor for Kpr modifications, and it is a product of cholesterol metabolism, odd-chain fatty acid oxidation, and branched-chain amino acid degradation. This suggests that Kpr may play a role in cellular metabolic regulation during fasting and catabolic processes by influencing chromatin structure.
Although the role of propionylation in cancer development has not been extensively studied, it is plausible that, given the synergistic effects of other acylation modifications in tumor pathology, propionylation may also play a significant role in tumor biology. These acylation modifications can affect protein stability, subcellular localization, and functionality, potentially contributing to tumor initiation and progression.
Protein functionality is regulated by various factors, including expression levels and post-translational modification states. Focusing solely on one aspect may overlook other critical elements. Therefore, to gain a comprehensive understanding of the trends in protein changes and the mechanisms of functional regulation, it is essential to conduct integrated analyses of quantitative proteomics and post-translational modification (PTM) data. Such analyses can provide systematic insights into changes at the protein level and enable a deeper exploration of protein function regulation. If you're interested, please refer to our other blog post that guides you on how to perform association analyses between quantitative proteomics and post-translational modification proteomics.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


