Exciting Upgrade: Expanded Database for Plant Widely Targeted Metabolomics
In the realm of plant metabolomics, the Plant Widely Targeted Metabolomics (WTP) service, patented by MetwareBio, has emerged as a powerful tool for comprehensive metabolite profiling. This unique platform combines the advantages of both targeted and untargeted metabolomics, offering a broad yet accurate analysis of plant metabolites. With its capacity to identify a vast range of plant metabolites with both precision and breadth, WTP has become an indispensable approach in plant biology research. Recently, we are thrilled to announce a significant upgrade to the WTP service's database, marking a new milestone in our commitment to providing cutting-edge metabolomics solutions.
What is Plant Widely Targeted Metabolomics?
Plant widely targeted metabolomics is an innovative approach that merges the strengths of untargeted and targeted metabolomics. Untargeted metabolomics enables the detection of a wide range of metabolites without prior knowledge, offering a holistic overview of the metabolome and uncovering novel compounds. However, its exploratory nature can sometimes compromise precision and quantification accuracy. On the other hand, targeted metabolomics focuses on specific known metabolites with high quantification accuracy and reproducibility, offering more reliable data on selected compounds. By integrating both strategies, WTP achieves comprehensive coverage with enhanced accuracy, making it an ideal solution for plant metabolomics research. This hybrid approach allows researchers to gain both broad insights and precise quantitative information, facilitating the discovery of novel metabolites alongside detailed quantification of known compounds.
 assay_1741139425_WNo_600d600.webp)
The workflow of Plant Widely Targeted Metabolomics (WTP) assay
Real-World Applications: Empowering Crop Improvement Research
One notable example of the WTP service's application is its role in a recent study published in Cell, where researchers combined evolutionary biology, genome editing, and metabolomics to engineer CoQ10-producing rice and wheat. By tracing CoQ side-chain variations across 134 plant species, the study revealed CoQ10 as the ancestral trait in flowering plants. The WTP service played a critical role in comprehensively profiling plant metabolites to assess the metabolic effects of introducing the CoQ10 biosynthetic pathway. The results demonstrated that the metabolic profiles of genetically modified plants showed no significant deviations from their wild-type counterparts, suggesting that CoQ10 biofortification does not induce unintended metabolic alterations. This finding supports the safety and viability of genetically engineered crops for human consumption and environmental sustainability, highlighting the indispensable role of WTP in risk assessment and crop improvement research.
 and in brown grain (C) of T1 edited Kitaake rice plants grown in green house._1741145403_WNo_938d319.webp)
 and metabolomics data (N, n = 5) from 20-DPA grains revealing very few differences between line 120 and _1741145104_WNo_708d374.webp)
CoQ9 and CoQ10 contents in 4-week-old leaf (B) and in brown grain (C) of T1 edited Kitaake rice plants grown in green house. Transcriptomics (M, n = 3) and metabolomics data (N, n = 5) from 20-DPA grains revealing very few differences between line 120 and Kitaake.
Expanded Database: Unlocking Greater Metabolite Diversity
The recent upgrade significantly expands the WTP database, which now includes over 61,000 plant metabolites—a remarkable leap from the previous 30,000+ metabolites. This expanded coverage encompasses a wide variety of plant metabolite classes, including 4,000+ primary metabolites essential for plant growth and development, as well as 56,000+ secondary metabolites such as lipids, flavonoids, alkaloids, phenylpropanoids, terpenoids, and more. Secondary metabolites play pivotal roles in plant defense mechanisms, signaling pathways, and interactions with the environment, making them key targets in plant metabolomics research. The upgraded database allows researchers to delve deeper into these complex biochemical networks, uncovering novel insights into plant physiology and adaptation.
Table of metabolites included in the database
|
Metabolite category |
Num. |
Representative compounds |
|
Flavonoids |
7800+ |
Rutin, Phloretin, Phelligrin A, Hesperetin, Licorice glycoside A, Pelargonidin-3-O-glucoside, Ginkgetin, Formononetin, Theaflavin |
|
Phenolic acids |
2500+ |
Chlorogenic acid, Momordicoside A, Oleuropein, Salvianolic acid A, Tatariside A, Veratric acid, Salidroside, Parishin B, Magnoloside A, Vanillic acid |
|
Alkaloids |
8500+ |
α-Solanine, Verticine, Nuciferine, Stachydrine, Matrine, Camptothecin, Arecoline, DIMBOA, Avenanthramide A, Lycorenine |
|
Terpenoids |
12000+ |
Artemisinine, Genipin, Paclitaxel, Wilforlide A, Protopanaxdiol, Saikosaponin A, Cucurbitacin B, Crocin I, Cyclocarioside Ⅰ, Ecliptasaponin A |
|
Quinones |
1000+ |
Emodin, Obtusin, Lapachone, Shikonin, Tectograndone, Morindaparvin A, Aloesin, 5-Hydroxydigitolutein, Trijuganone A |
|
Steroid |
1600+ |
Asparagoside C, Polyphyllin I, Timosaponin A-III, Gracillin, Sarsasapogenin, Tigogenin, Digitonin, Oleandrin |
|
Tannins |
400+ |
Ellagic acid, Gemin D, Casuariin, Punicalin, Chebulagic acid, 1,3,6-Tri-O-galloylglucose, Chebulanin, Tellimagrandin I |
|
Lignans |
1100+ |
Honokiol, Syringaresinol, Arctigenin, Pinoresinol, Schisanhenol, Sesamin, Chestnutlignansoide, Trachelogenin, Fargesin, Isolariciresinol |
|
Glucosinolates |
200+ |
Sulforaphane, Gluconasturtiin, Sinalbin, Glucocheirolin, Glucoraphanin, 4-Hydroxy-3-indolylmethyl glucosinolate, Sinigrin, 4-Methylsulfinyl-3-Butenyl Glucosinolate |
|
Coumarins |
1500+ |
Umbelliprenin, Psoralen, Glycycoumarin, Xanthotoxol, Scopolin, Bengenin, Bergapten, Decursinol, Dihydrocoumarin |
|
2100+ |
Succinic acid, Malic acid, Citric Acid, Quinic Acid, Abscisic acid, Tartaric acid, Shikimic acid, Aconitic Acid, Salicylic acid, Cinnamic acid, Maleic acid |
|
|
Vitamins |
50+ |
Vitamin C, Vitamin B2, Vitamin A1, Vitamin U, Ginkgotoxin, Nicotinic acid, Nicotinamide, Retinol, Vitamin D3, Tocotrienol |
|
800+ |
Tryptophan, Theanine, Beauvericin, Dencichin, Heterophyllin, Saccharopine, Alliin, Dopa, S-Adenosylmethionine, γ-Glu-Cys |
|
|
Nucleotides and derivatives |
300+ |
Adenine, Cytosine, Thymine, Inosine, Eritadenine, Xanthosine, Cordycepin, Sepiapterin, Adenosine 5'-monophosphate |
|
Saccharides and Alcohols |
450+ |
Glucose, Fructose, Sucrose, Fucose, Xylitol, Rhamnose, Maltose, Raffinose, Allitol, Mannitol |
|
Lipids |
900+ |
Linolenic acid, 4-Hydroxysphinganine, Lauric acid, Myristic Acid, Palmitic acid, Arachidonic Acid, Stearic Acid |
|
Others |
20000+ |
Aflatoxin B1, Secoxyloganin, Kavain, Terreic acid, Mansonone E, Litchiol A, Myricanol, Safranal, Bruceine A, Gambogic acid |
|
Total |
61150+ |
|
Unveiling Unprecedented Detection Capability with the New Database
With this upgraded database, the number of metabolites detected through the WTP service has significantly increased, enhancing the overall detection rate across various plant species and sample types. This expanded detection capacity is particularly valuable for researchers studying plant secondary metabolism, where even subtle metabolic variations can have profound biological significance. Moreover, the number of metabolites identified at qualitative level 1—the highest confidence level based on standard reference materials—has notably increased. This improvement ensures greater accuracy and reliability in metabolite annotation, providing researchers with more robust data for downstream analyses such as pathway reconstruction, biomarker discovery, and comparative metabolomics.
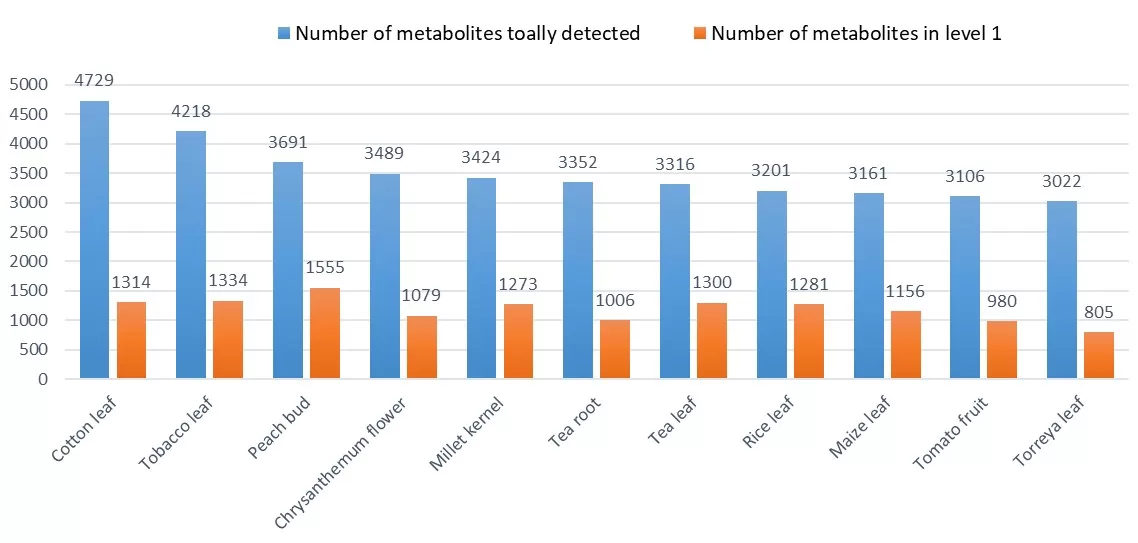
Past experience on various plant samples across different species
How Can This Upgrade Transform Your Plant Research?
This upgrade empowers researchers with deeper insights into plant metabolism, particularly in areas such as plant stress responses, metabolite biosynthesis pathways, and plant-environment interactions. The expanded database will support a wider range of research applications, from functional genomics and natural product discovery to crop improvement and ecological studies. Whether unraveling the metabolic basis of stress tolerance, identifying novel bioactive compounds, or enhancing crop nutritional value, the upgraded WTP service offers an unparalleled resource for plant biology research.
With the continuous evolution of our plant widely targeted metabolomics service, we remain dedicated to providing researchers with advanced analytical tools that push the boundaries of metabolomics research. The expanded database not only enhances metabolite coverage and detection rates but also reinforces our commitment to delivering high-quality, reproducible data to the global plant research community.
Explore the enhanced capabilities of our upgraded WTP service and unlock new possibilities for your plant metabolomics studies!
Reference
Xu JJ, Lei Y, Zhang XF, Li JX, Lin Q, Wu XD, Jiang YG, Zhang W, Qian R, Xiong SY, Tan K, Jia Y, Zhou Q, Jiang Y, Fan H, Huang YB, Wang LJ, Liu JY, Kong Y, Zhao Q, Yang L, Liu J, Hu YH, Zhan S, Gao C, Chen XY. Design of CoQ10 crops based on evolutionary history. Cell. 2025 Feb 11:S0092-8674(25)00087-X. doi: 10.1016/j.cell.2025.01.023.


