Key Methods in Plant Proteomics: Protein Extraction Techniques
Introduction to Plant Proteomics and Its Importance
Plant samples occupy a pivotal position in proteomics research due to their vast biochemical diversity and significance in various fields, including agriculture, nutrition, and environmental science. The increasing focus on plant proteomics is driven by the need to improve crop yield, enhance nutritional content, and develop sustainable agricultural practices, underscoring the importance of integrating proteomic analysis with genomics and metabolomics to fully elucidate the molecular mechanisms underlying plant biology. As methodologies continue to evolve, the ability to extract and analyze proteins from diverse plant samples will further enhance our understanding of plant systems and their applications in biotechnology and ecological research.
Limitations and Challenges in Plant Protein Extraction
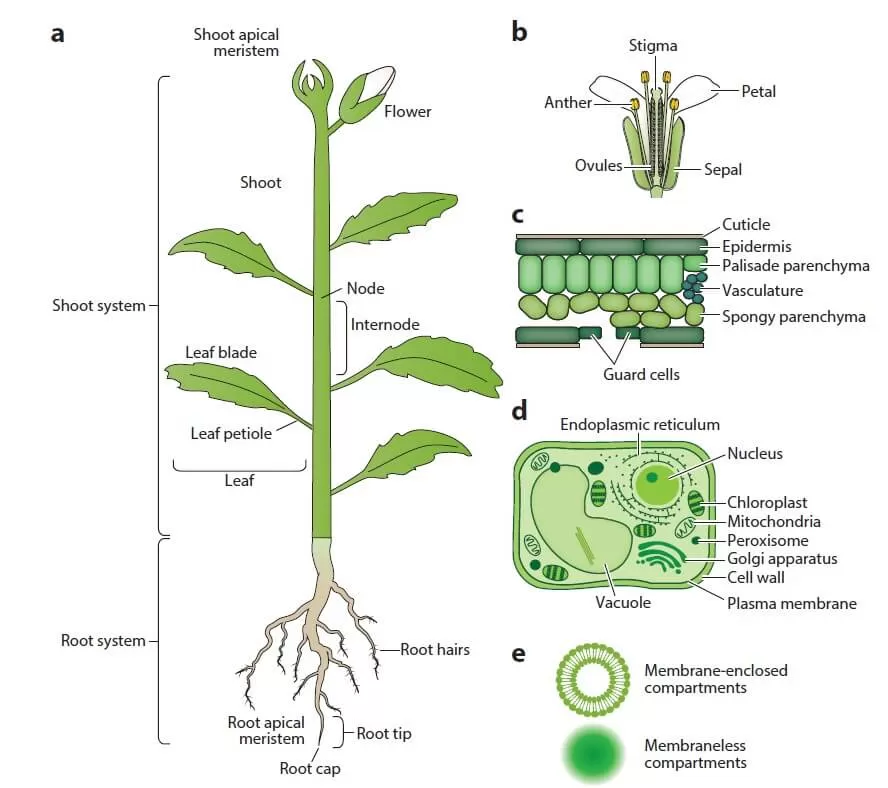
Unlike most mammals, plants are considered structurally simpler, with a limited number of organs (see Figure 1). These are categorized into two types: vegetative organs (roots, stems, and leaves) and reproductive organs (flowers, fruits, and seeds). A typical plant body consists of two systems:
- Root system (below ground): Anchors the plant in the soil, absorbs water and minerals, and stores some products of photosynthesis.
- Shoot system (above ground): Includes stems that transport water and minerals from the roots to leaves, where photosynthesis occurs, generating energy and synthesizing organic compounds. During reproduction, the shoot produces flowers, which after fertilization develop into fruits that carry seeds.
Though plant structures are simpler, protein extraction from plant tissues poses more challenges compared to human or animal samples. Plant cells have a lower protein yield (protein abundance) because a significant portion of the cell is occupied by the cell wall and vacuole. The two most common methods for breaking down tough plant cell walls are grinding frozen plant material in liquid nitrogen using a mortar and pestle (low throughput) or bead milling (medium throughput). Keeping plant material frozen during cell disruption helps minimize protein degradation and unwanted modifications. The cell wall and vacuole contain numerous compounds that interfere with protein sample preparation, including phenolic compounds, proteases, oxidases, terpenes, pigments, and carbohydrates. The composition of these primary and secondary metabolites varies widely across species, organs, and developmental stages. For instance, mature green tissues are rich in secondary metabolites, while rapidly dividing meristematic cells contain higher levels of nucleic acids. Additionally, certain tissues, such as leaves and seeds, are dominated by storage proteins.
The ideal protein extraction method should reliably capture all proteins with minimal contamination from other molecules. However, due to the diversity in protein abundance, molecular weight, charge, hydrophobicity, post-translational modifications, and complex formations, no single extraction method works for all proteins. Thus, sample preparation is a critical step in plant proteomics research.
Mainstream Methods for Plant Protein Extraction and Their Advantages
Two classic methods for extracting plant proteins are the trichloroacetic acid (TCA)-acetone precipitation method and the phenol extraction method.
The TCA/acetone precipitation method has been used since the 1980s for protein separation in 1D and 2D-PAGE analyses. This extraction process involves denaturing and precipitating proteins, with the addition of 2-mercaptoethanol (2-ME) in the buffer to inhibit the formation of new disulfide bonds. Most interfering compounds remain soluble in acetone, which is removed, leaving behind a granular protein precipitate. While this denaturation under acidic and/or hydrophobic conditions aids in protein concentration and impurity removal, it has some drawbacks: (1) the resulting granules may be difficult to dissolve; (2) TCA can precipitate nucleic acids longer than 20 nucleotides; and (3) in some cases, TCA may hydrolyze proteins.
The phenol extraction method, another common technique, involves dissolving proteins in phenol and then precipitating them with methanol and ammonium acetate. This method has been shown to yield high-quality proteins from various plant species. Commonly used by molecular biologists for nucleic acid extraction, this method is beneficial for extracting plant tissue proteins due to its ability to remove interfering compounds as well as DNA.
-acetone precipitation method and phenol extraction method for th_1727420594_WNo_242d246.webp) In the phenol extraction method, the first step involves mixing a tris(hydroxymethyl)aminomethane (Tris) buffer with a sucrose buffer to separate proteins from salts, nucleic acids, and carbohydrates. This phase separation allows the phenol phase to be placed on top. Denatured proteins dissolved in the phenol phase are then precipitated with methanol and ammonium acetate, followed by washing with acetone and methanol to remove pigments and lipids. However, this method can be time-consuming, and the resulting particles may be difficult to dissolve. Key considerations include:
In the phenol extraction method, the first step involves mixing a tris(hydroxymethyl)aminomethane (Tris) buffer with a sucrose buffer to separate proteins from salts, nucleic acids, and carbohydrates. This phase separation allows the phenol phase to be placed on top. Denatured proteins dissolved in the phenol phase are then precipitated with methanol and ammonium acetate, followed by washing with acetone and methanol to remove pigments and lipids. However, this method can be time-consuming, and the resulting particles may be difficult to dissolve. Key considerations include:
- The phenol extraction method is more effective for dissolving membrane-associated proteins, whereas the TCA/acetone method is better suited for water-soluble proteins.
- Using protease inhibitors is not essential for either method, as the acidic or organic reagents used denature proteins and inhibit protease activity, minimizing protein degradation during extraction without heating.
- While manual grinding of tissues can enhance protein yield, automated bead milling with grinding balls and a vibrating homogenizer can also be employed to increase yield and improve reproducibility across different samples.
- The efficiency of protein dissolution from pellets after phenol extraction is higher compared to the TCA/acetone method, reducing protein loss and resulting in higher purity. Despite these advantages, the phenol extraction method has limitations, as it may result in the loss of specific proteins, particularly large glycoproteins.
._1727430281_WNo_294d287.webp) Comparison of the TCA (Trichloroacetic Acid)-acetone precipitation method and phenol extraction method for the same sample. (a) Fine tissue powder obtained by grinding in a mortar; (b) Tissue powder thoroughly homogenized in TCA/acetone in the mortar; (c) Tissue pellets (white or light-colored) in an EP tube after washing with TCA/acetone; (d) Phase separation during phenol extraction, with the phenolic phase containing proteins forming a lower phase in the EP tube; (e) Protein precipitation with ammonium acetate in methanol.
Comparison of the TCA (Trichloroacetic Acid)-acetone precipitation method and phenol extraction method for the same sample. (a) Fine tissue powder obtained by grinding in a mortar; (b) Tissue powder thoroughly homogenized in TCA/acetone in the mortar; (c) Tissue pellets (white or light-colored) in an EP tube after washing with TCA/acetone; (d) Phase separation during phenol extraction, with the phenolic phase containing proteins forming a lower phase in the EP tube; (e) Protein precipitation with ammonium acetate in methanol.
Advancements in Plant Protein Extraction Techniques
In the sample preparation workflow, combining protein precipitation methods such as trichloroacetic acid (TCA)/acetone, phenol, or chloroform with metabolite extraction can help remove substances that may interfere with protein solubility, peptide digestion, and the purification process prior to LC-MS/MS analysis. However, the drawback of protein precipitation methods is their relatively low solubility efficiency, often leading to significant sample loss. Variability in the sample preparation process can also complicate quantitative comparisons. As proteomics research advances and the diversity of protein types increases, various strategies have been developed to enhance the coverage and detection of specific proteomes, such as membrane proteins and low-abundance proteins. These strategies include:
- Sequential extraction using a series of reagents based on different protein solubilities.
- Phase separation using organic solvents or detergents like Triton X-114.
- Liquid chromatography (LC) fractionation.
- Isolation of highly enriched organelles or subcellular compartments, including chloroplasts, mitochondria, nuclei, vacuoles, peroxisomes, microsomes, plasma membranes, cell walls, and extracellular matrices.
These front-end fractionation procedures significantly enhance detection capabilities and resolution while reducing overall sample complexity, thus broadening the protein coverage, including low-abundance proteins.
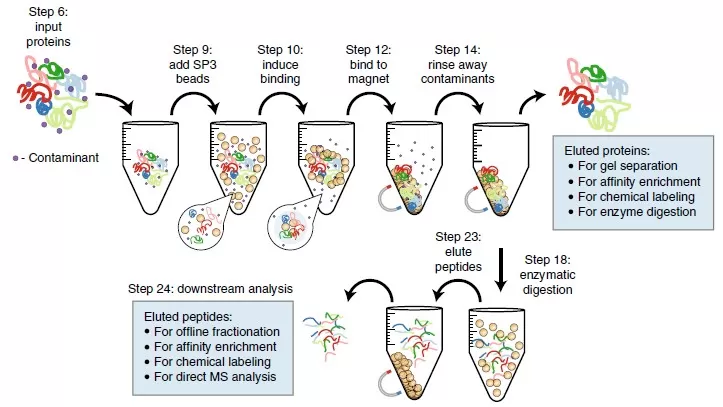
Some methods initially developed for mammalian systems, which use strong detergents or chaotropes to dissolve or precipitate proteins from cells, can also be applied to plant protein extraction. These methods include Filter-Aided Sample Preparation (FASP), S-Trap Protein Suspension Capture, Gel-Lysis Protein Digestion (GeLC-MS), and Single-Pot Solid-Phase Enhanced Sample Preparation (SP3). They utilize surfactants such as sodium dodecyl sulfate (SDS), 3-((3-cholamidopropyl)dimethylammonio)-1-propanesulfonate (CHAPS), sodium deoxycholate (SDC), chaotropes (urea, thiourea), reducing agents (such as dithiothreitol, DTT), or combinations of various reagents to maximize protein solubility.
For instance, SDS is an effective solubilizing and denaturing agent that allows for the analysis of even multi-pass membrane proteins. However, the use of SDS in upstream LC-MS/MS analysis has long been problematic, as even trace amounts can obscure peptide signals. The recently developed SP3 method elegantly addresses this issue by precipitating proteins onto microspheres and utilizing high organic content solvents to keep SDS in solution. Subsequent digestion of proteins on the beads enables scalability to a 96-well format, enhancing throughput and reproducibility while allowing for very small input amounts, thus improving sensitivity. This approach, extracting proteins using SDT extraction buffer and then utilizing SP3 to remove interfering substances like SDS before enzymatic digestion on microspheres, has been successfully applied to Arabidopsis leaf tissues. It demonstrates significant potential for high-throughput plant proteomics, serving as a reproducible method for sample preparation across various tissue types and plant species.
Reference
1. Mergner J, Kuster B. Plant Proteome Dynamics. Annu Rev Plant Biol. 2022 May 20;73:67-92. doi: 10.1146/annurev-arplant-102620-031308. Epub 2022 Feb 9. PMID: 35138880.
2. Alvarez S, Naldrett MJ. Plant Structure and Specificity - Challenges and Sample Preparation Considerations for Proteomics. Adv Exp Med Biol. 2016;919:63-81. doi: 10.1007/978-3-319-41448-5_4. PMID: 27975213.
3. Wu X, Xiong E, Wang W, Scali M, Cresti M. Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat Protoc. 2014 Feb;9(2):362-74. doi: 10.1038/nprot.2014.022. Epub 2014 Jan 16. PMID: 24434803.
4. Al-Mohanna T, Bokros NT, Ahsan N, Popescu GV, Popescu SC. Methods for Optimization of Protein Extraction and Proteogenomic Mapping in Sweet Potato. Methods Mol Biol. 2020;2139:309-324. doi: 10.1007/978-1-0716-0528-8_23. PMID: 32462596.
5. Galazzi RM, de Jesus JR, Arruda MAZ. Sample Preparation Focusing on Plant Omics. Adv Exp Med Biol. 2019;1073:161-185. doi: 10.1007/978-3-030-12298-0_7. PMID: 31236843.
6. Hughes CS, Moggridge S, Müller T, Sorensen PH, Morin GB, Krijgsveld J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc. 2019 Jan;14(1):68-85. doi: 10.1038/s41596-018-0082-x. PMID: 30464214.
7. Mikulášek K, Konečná H, Potěšil D, Holánková R, Havliš J, Zdráhal Z. SP3 Protocol for Proteomic Plant Sample Preparation Prior LC-MS/MS. Front Plant Sci. 2021 Mar 10;12:635550. doi: 10.3389/fpls.2021.635550. PMID: 33777071; PMCID: PMC7988192.
._1727420537_WNo_323d317.webp)
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


