Phosphoproteomics in Action: Strategies for Investigating Protein Phosphorylation
Phospho-proteomics is a scientific discipline that studies protein phosphorylation modifications. It reveals protein functions and regulatory networks by identifying and quantifying phosphorylation sites on proteins. Protein phosphorylation is a major post-translational modification (PTM) where a phosphate group from ATP is transferred to specific amino acid residues. In eukaryotic cells, around 30% of proteins undergo phosphorylation, which is crucial for understanding cellular functions.
Mechanism of Protein Phosphorylation
Phosphorylation involves two key reversible steps, regulated by protein kinases and phosphatases:
1. Phosphorylation: A kinase transfers a phosphate group from ATP to a target protein's amino acid residue, typically serine, threonine, or tyrosine, modifying the protein's structure and function.
2. Dephosphorylation: A phosphatase removes the phosphate group, restoring the protein’s original state. This balance is essential for proper cellular signaling.
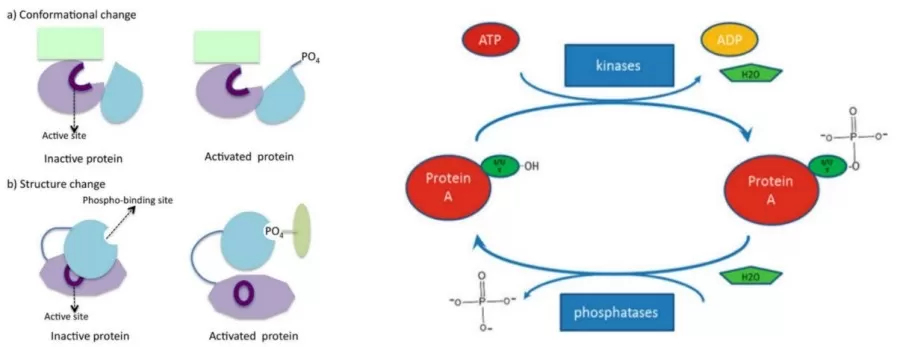
Both processes are tightly regulated to maintain accurate signal transduction, with each modification impacting protein activity, localization, and protein-protein interactions, affecting key processes like cell growth, division, and apoptosis.
Types of Protein Phosphorylation
Phosphorylation is classified based on the modified amino acid residue:
1. Serine Phosphorylation: The most common type, involved in regulating signal transduction, protein interactions, and enzymatic activity by altering protein conformation.
2. Threonine Phosphorylation: Similar to serine, threonine phosphorylation modulates cell signaling and metabolic processes.
3. Tyrosine Phosphorylation: This is pivotal in eukaryotic cell signaling, particularly in cell proliferation, differentiation, and apoptosis. It also acts as a signaling molecule to form complexes with other proteins, propagating signals.
Though rarer, phosphorylation can also occur on histidine, lysine, and arginine residues, but their specific functions are still under investigation.
Common Methods Used in Phospho-Proteomics Research
1. Mass Spectrometry (MS)
Mass spectrometry is a high-resolution analytical technique for studying protein phosphorylation. It measures the mass-to-charge ratio of peptides, allowing for the identification and quantification of phosphorylation sites with precision.
2. Western Blotting
Western blotting involves separating proteins by gel electrophoresis, transferring them onto a membrane, and then detecting specific phosphorylation sites using antibodies. Only phosphorylated proteins will yield distinct bands at their corresponding molecular weights, allowing for targeted analysis.
3. Immunoprecipitation (IP)
Immunoprecipitation employs specific antibodies to isolate target proteins from cell lysates. The precipitated proteins can then be analyzed for phosphorylation status using techniques such as Western blotting or other detection methods.
4. Protein Phosphorylation Arrays
Protein phosphorylation arrays are high-throughput platforms that enable the simultaneous detection of phosphorylation states for multiple proteins in a single experiment. Specific antibodies are immobilized on the array and bind to phosphorylated proteins in the sample, which are then detected using fluorescence or chemiluminescence.
5. Flow Cytometry
Flow cytometry characterizes single cells by utilizing fluorescently labeled antibodies that target specific phosphorylated proteins. The average fluorescence intensity measured reflects the phosphorylation levels of the target proteins, providing quantitative insights into cellular signaling dynamics.
These methods collectively enhance our understanding of protein phosphorylation, contributing to insights into cellular functions and regulatory mechanisms.
Methods and Workflow of MS-Based Phospho-Proteomics
1. Sample Preparation
Choose appropriate sample types, including animal tissues, cells, plant samples, and body fluids, ensuring their representativeness and accuracy. During the processing phase, it is crucial to handle the samples promptly to prevent protein degradation. The use of protease and phosphatase inhibitors is recommended to preserve protein integrity.
2. Sample Pre-Treatment
Extract total proteins from the selected samples and digest them into peptides to facilitate subsequent analysis.
3. Enrichment of Phosphorylated Peptides
Prior to mass spectrometry analysis, phosphorylated peptides typically require enrichment. Common enrichment methods include:
- Metal Oxide Affinity Chromatography (MOAC): For example, titanium dioxide (TiO₂) enrichment utilizes the ability of metal oxides to bind with oxygen in phosphate groups, effectively capturing phosphorylated peptides.
- Immunoprecipitation: This technique specifically enriches tyrosine-phosphorylated peptides, allowing for high specificity in targeting phosphorylation sites.
-
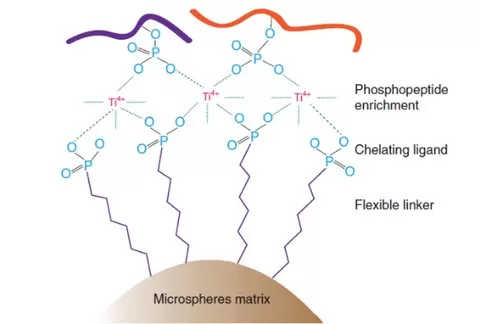 Immobilized Metal Ion Affinity Chromatography (IMAC): This method employs transition metal cations, such as Fe³⁺, Ga³⁺, and Zr⁴⁺, as affinity reagents to interact with anionic phosphorylated peptides. The next-generation Ti-IMAC technology uses tetravalent titanium ions (Ti⁴⁺) to achieve specific capture of phosphorylated proteins or peptides through electrostatic interactions with phosphate groups. Under optimal buffer conditions, phosphorylated proteins or peptides bind to Ti⁴⁺ ions, while non-phosphorylated proteins remain unbound, facilitating the effective enrichment of phosphorylated proteins.
Immobilized Metal Ion Affinity Chromatography (IMAC): This method employs transition metal cations, such as Fe³⁺, Ga³⁺, and Zr⁴⁺, as affinity reagents to interact with anionic phosphorylated peptides. The next-generation Ti-IMAC technology uses tetravalent titanium ions (Ti⁴⁺) to achieve specific capture of phosphorylated proteins or peptides through electrostatic interactions with phosphate groups. Under optimal buffer conditions, phosphorylated proteins or peptides bind to Ti⁴⁺ ions, while non-phosphorylated proteins remain unbound, facilitating the effective enrichment of phosphorylated proteins.
Advantages of Ti-IMAC Enrichment:
- High Specificity: Ti-IMAC effectively recognizes and binds phosphorylated proteins or peptides, minimizing interference from non-specific interactions.
- High Enrichment Efficiency: Compared to traditional phosphorylation enrichment methods, Ti-IMAC demonstrates significantly higher efficiency, leading to a notable increase in the number of identified phosphorylated proteins.
- Broad Applicability: This method is suitable for enriching phosphorylated proteins from a variety of biological samples, including mammalian cells, tissues, bacteria, yeast, and plants.
4. Mass Spectrometry Analysis
Mass spectrometry is a pivotal tool in phospho-proteomics research, allowing for the identification and quantification of phosphorylated peptides. Electrospray ionization mass spectrometry (ESI-MS) is widely utilized, where the sample is mixed with protonation agents and injected into the mass spectrometer. This technique facilitates the analysis of specific phosphorylation sites and levels. Common mass spectrometry scanning modes include:
- Data-Dependent Acquisition (DDA): Gathers data based on previously detected ions.
- Data-Independent Acquisition (DIA): Collects data from all ions within a defined range, enhancing data completeness and reliability.
5. Data Analysis
The data obtained from mass spectrometry must be analyzed using specialized software to identify phosphorylation sites and quantify their levels. Commonly used software includes MaxQuant, Proteome Discoverer, and Spectronaut, which aid in the interpretation of complex datasets.
6. Bioinformatics Analysis
Identified phosphorylation sites can be subjected to further bioinformatics analysis, including Gene Ontology (GO) analysis, pathway analysis, and protein-protein interaction network predictions. These analyses provide insights into the biological roles of phosphorylation events in various cellular processes.
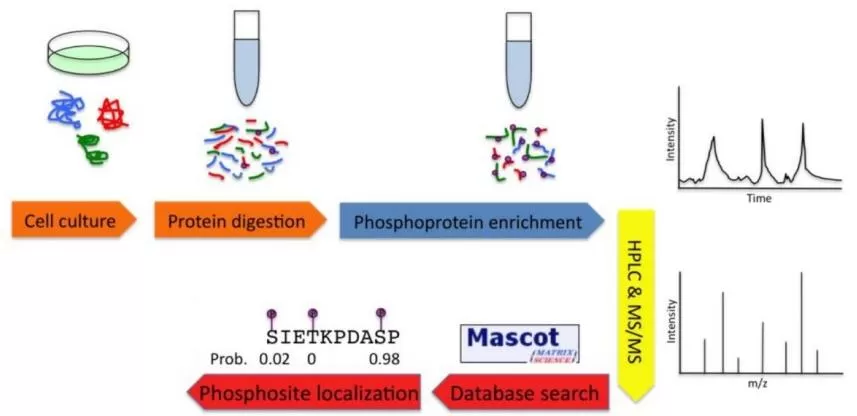
By implementing these methods and workflows, researchers can gain significant insights into the dynamics of protein phosphorylation, contributing to a deeper understanding of cellular signaling, regulation, and the biological significance of phosphorylation in diverse biological systems.
Sample Types for MS-Based Phospho-Proteomics
Various sample types can be utilized for phospho-proteomics analysis. Here are some common types suitable for studying protein phosphorylation modifications:
1. Animal Tissues
Tissues such as heart, liver, spleen, lung, kidney, intestine, brain, and muscle can be used. After washing with phosphate-buffered saline (PBS) to remove residual blood and contaminants, these tissues should be snap-frozen in liquid nitrogen and stored at -80°C for subsequent analysis.
2. Plant Samples
Samples from stems, leaves, flowers, roots, bark, and branches of both woody and herbaceous plants, as well as algae and ferns, can be included. After thorough washing, these samples should also be snap-frozen in liquid nitrogen for preservation.
3. Microbial Samples
Bacterial and fungal samples can be collected and must undergo centrifugation, washing, and snap-freezing in liquid nitrogen to preserve their phosphorylation states.
4. Cell Samples
Both suspended and adherent cultured cells are suitable. After collection, cells should be centrifuged to remove the culture medium and then snap-frozen in liquid nitrogen.
5. Body Fluid Samples
Plasma and serum samples are valuable for phospho-proteomics. Following collection, these samples should be centrifuged to separate serum or plasma, treated with protease inhibitors, and snap-frozen in liquid nitrogen.
Overall, a wide range of sample types is applicable in phospho-proteomics research, each requiring specific handling protocols to ensure the accuracy and reliability of experimental results.
Sample Preparation Considerations for MS-Based Phospho-Proteomics
During sample preparation, the following key points should be considered:
1. Ensure Sample Representativeness and Accuracy
Select samples directly relevant to the research question, such as specific tissues, cell types, or biological fluids. The collection and handling of samples should accurately reflect the research objectives.
2. Maintain Sample Consistency and Timeliness
Minimize variability among replicate samples and reduce the time from collection to experimentation to ensure reproducibility.
3. Preserve Samples at Low Temperatures
Keep samples at low temperatures during processing and storage to prevent metabolic changes and alterations in phosphorylation status.
4. Detail Special Treatment Conditions
For samples subjected to drug treatment or stress conditions, document these treatments thoroughly to inform subsequent analyses.
5. Protect Phosphorylation States During Extraction and Detection
Use phosphatase and protease inhibitors during the extraction and detection of phosphorylated proteins, and avoid high temperatures or prolonged processing that could alter phosphorylation states.
6. Conduct Pilot Experiments
Perform preliminary experiments to determine optimal induction conditions and timing, ensuring adequate expression and modification levels of phosphorylated proteins.
7. Utilize Databases for Phosphorylation Information
Leverage databases like PhosphoSitePlus to query phosphorylation status and incorporate experimental design and pilot studies to assess phosphorylation expression levels in samples.
Choosing the appropriate sample types and attending to the details of sample handling will enhance the success rate and reliability of phospho-proteomics studies.
Challenges and Future Directions of MS-Based Phospho-Proteomics
1. Sample Processing Difficulties
The transient and reversible nature of phosphorylation imposes strict requirements on sample collection and preservation. Moreover, the negative charge introduced by phosphate groups can complicate the ionization of modified peptides during analysis.
2. Development of New Technologies
Future research will focus on developing innovative enrichment strategies and mass spectrometry techniques to enhance the sensitivity and accuracy of phospho-proteomics. For example, integrating magnetic Fe3O4 and TiO2 particle-based sequential enrichment techniques shows promise for improving the quality of experimental data.
By addressing these challenges and pursuing advancements in technology, the field of phospho-proteomics can continue to grow, providing deeper insights into cellular signaling and regulation.
Reference
1. Qu L,Liu M,Zheng L, et al. Data-independent acquisition-based global phosphoproteomics reveal the diverse roles of casein kinase 1 in plant development. Sci Bull (Beijing). 2023;68 (18):2077-2093. doi:10.1016/j.scib.2023.08.017
2. Zhao M,Cao N,Gu H, et al. AMPK Attenuation of β-Adrenergic Receptor-Induced Cardiac Injury via Phosphorylation of β-Arrestin-1-ser330. Circ Res. 2024;135 (6):651-667. doi:10.1161/CIRCRESAHA.124.324762
3. Maďarová M,Mucha R,Hresko S, et al. Identification of new phosphorylation sites of CD23 in B-cells of patients with chronic lymphocytic leukemia. Leuk Res. 2018;70:25-33. doi:10.1016/j.leukres.2018.05.002
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


