Decoding Peptide Mass Fingerprinting: A Guide to Protein Identification in Proteomics
In the realm of biological sciences, proteins play a crucial role. They are the fundamental building blocks of living organisms and are involved in various physiological processes, such as metabolism, signal transduction, and cellular behavior. However, due to the vast diversity of proteins, traditional identification techniques often fall short of providing a comprehensive understanding of the types and functions of proteins in complex biological systems. Fortunately, Peptide Mass Fingerprinting (PMF) offers a new perspective, enabling us to identify and study proteins with unprecedented accuracy.
Peptide Mass Fingerprinting is a powerful protein identification technique that determines the identity of proteins by analyzing the mass of peptides generated through protein digestion. As an advanced and robust detection method, what are its principles, features, and applications? Join us as we unveil the mysteries of Peptide Mass Fingerprinting and explore this fascinating world of biological science!
Principle of Peptide Mass Fingerprinting (PMF)
The fundamental principle of Peptide Mass Fingerprinting (PMF) involves using specific proteases, such as trypsin, to cleave proteins into smaller peptide fragments. These fragments are then subjected to precise mass measurement using mass spectrometry techniques, such as MALDI-TOF or ESI-TOF. Each protein has a unique amino acid sequence, leading to distinct peptide fragments after enzymatic digestion, which collectively create a unique "fingerprint" for that protein. By comparing the experimentally obtained peptide mass data with known protein fingerprints in a database, the identity of an unknown protein can be determined.
Workflow of Peptide Mass Fingerprinting (PMF)
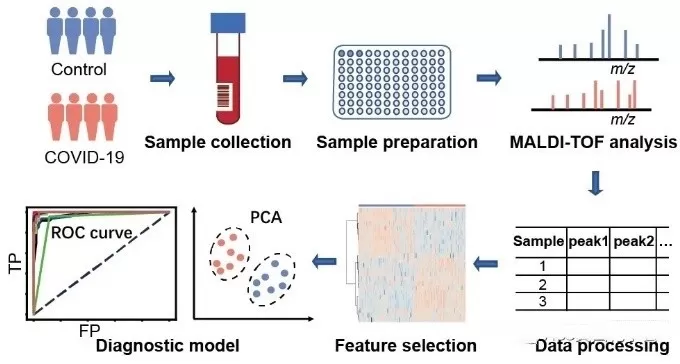
The workflow of Peptide Mass Fingerprinting (PMF) typically consists of several key steps, each crucial for accurately identifying proteins based on their peptide mass profiles. Here’s a detailed overview of the process:
1. Sample Preparation:
Protein Extraction: The first step involves extracting proteins from biological samples, such as cells, tissues, or biofluids, using appropriate lysis buffers and methods to preserve protein integrity.
Protein Quantification: The concentration of the extracted proteins is measured using methods like the Bradford assay or BCA assay to ensure sufficient amounts for analysis.
2. Enzymatic Digestion:
Proteolytic Digestion: The extracted proteins are digested into smaller peptide fragments using specific proteases, with trypsin being the most commonly used. Trypsin cleaves proteins at specific sites (typically after lysine and arginine residues), resulting in a predictable set of peptide fragments.
Sample Cleanup: The digested samples may undergo purification steps, such as solid-phase extraction, to remove contaminants and salts that could interfere with mass spectrometry analysis.
3. Mass Spectrometry Analysis:
Ionization: The purified peptide mixture is ionized using a mass spectrometry technique, such as Matrix-Assisted Laser Desorption/Ionization (MALDI) or Electrospray Ionization (ESI). This process converts the peptides into charged ions for analysis.
Mass Measurement: The ionized peptides are introduced into the mass spectrometer, where their mass-to-charge ratios (m/z) are measured. This generates a mass spectrum that displays the different peptide masses.
4. Data Analysis:
Database Search: The resulting mass spectrum is compared to a database of known protein sequences and their theoretical peptide mass fingerprints. Algorithms match the experimental data to potential protein identities based on peptide mass and abundance.
Identification: By matching the experimental peptide masses with those in the database, the software can identify the protein(s) present in the original sample. This step may also provide information about protein modifications.
Characteristics of Peptide Mass Fingerprinting (PMF)
Peptide Mass Fingerprinting (PMF) is one of the most commonly used and effective identification methods in proteomics research. Compared to traditional protein identification techniques, PMF exhibits the following characteristics:
- High Specificity: Each protein generates unique peptide masses due to the distinct amino acid sequences. This uniqueness provides PMF with exceptional specificity, allowing for accurate protein identification.
- High Sensitivity: Advances in mass spectrometry technology have significantly enhanced the sensitivity of PMF, enabling the detection of proteins present at very low abundance.
- Good Stability: The results obtained from mass spectrometry analyses are stable and reliable, making them suitable for long-term storage and repeat verification.
- Minimal Sample Loss: Unlike traditional methods such as two-dimensional gel electrophoresis, mass spectrometry requires less sample material, thereby reducing sample loss during the analysis process.
Applications of Peptide Mass Fingerprinting (PMF)
With the advancement of analytical chemistry, the applications of Peptide Mass Fingerprinting (PMF) have become increasingly diverse, spanning from fundamental biological research to drug development and disease diagnosis.
1. Basic Biological Research
In fundamental biological research, PMF plays a crucial role in protein discovery and characterization. It helps scientists identify new proteins, elucidate their structures, and understand their functions and interactions within cellular pathways. For instance, PMF is instrumental in identifying post-translational modifications, such as phosphorylation and glycosylation, which can significantly impact protein function and activity. Furthermore, PMF aids in the characterization of protein complexes, enabling researchers to dissect protein-protein interactions and their biological significance.
2. Drug Development
In the realm of drug development, PMF is a valuable tool for identifying potential drug targets or biomarkers. By comparing the PMF profiles of proteins from diseased versus healthy tissues, researchers can pinpoint proteins that are differentially expressed, which may serve as promising targets for new therapies. Additionally, PMF can be employed in the early stages of drug screening to evaluate the efficacy of compounds on specific proteins, streamlining the drug discovery process. The technology also facilitates the validation of therapeutic targets by confirming the presence and abundance of proteins in various biological samples.
3. Disease Diagnosis
PMF offers clinicians a non-invasive method for identifying and analyzing protein differences in patient samples, which can be critical for early diagnosis and monitoring of diseases. For example, PMF can be used to detect protein biomarkers associated with cancer, cardiovascular diseases, and neurodegenerative disorders. This capability enables the development of diagnostic tests that can lead to personalized treatment strategies. Furthermore, PMF is utilized in monitoring disease progression and treatment response by tracking changes in protein expression over time, providing valuable insights into the effectiveness of therapeutic interventions.
4. Proteomics and Systems Biology
Beyond individual applications, PMF contributes significantly to the fields of proteomics and systems biology. It aids in constructing comprehensive protein interaction networks, providing insights into the complex interplay between proteins within biological systems. By integrating PMF data with genomic and transcriptomic information, researchers can gain a holistic understanding of cellular functions, pathways, and the molecular basis of diseases.
5. Biotechnology and Industry
PMF is also applied in biotechnological and industrial settings, where it helps in the quality control of biopharmaceutical products. By characterizing protein therapeutics, PMF ensures consistency and purity, which are critical for regulatory compliance and patient safety. Additionally, PMF is employed in enzyme engineering, where it assists in identifying and optimizing enzyme properties for industrial applications.
6. Environmental and Food Sciences
In environmental and food sciences, PMF can be utilized to identify proteins that serve as biomarkers for pollution or food quality. For example, PMF can aid in detecting specific proteins that indicate contamination in water sources or assess the quality of food products by profiling protein content and modifications.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


