Peptide and protein de novo sequencing by mass spectrometry (MS) is a cutting-edge technique that allows researchers to determine the amino acid sequence of proteins and peptides without relying on prior sequence information. This method has revolutionized proteomics, enabling the analysis of proteins from organisms with incomplete or unknown genomic data. The process begins with the enzymatic digestion of proteins into peptides, which are then analyzed by a mass spectrometer. The instrument generates precise mass-to-charge ratios (m/z) and fragment ions, which are further analyzed to deduce the peptide’s amino acid sequence. By using tandem mass spectrometry (MS/MS), researchers can gain deeper insights into the structural complexities of proteins, as the fragmentation patterns provide critical information for sequence determination.
The Basic Principle of Peptide and Protein De Novo Sequencing by Mass Spectrometry
Peptide and protein de novo sequencing by mass spectrometry (MS) operates on a fundamental principle of measuring the mass-to-charge (m/z) ratios of ions produced from peptide fragmentation. The core concept behind this method is the ability to deduce the amino acid sequence of peptides and proteins without prior knowledge of the sequence. Initially, proteins are enzymatically digested into smaller peptides, which are then introduced into the mass spectrometer. As peptides are ionized, they pass through a mass spectrometer where they are fragmented into smaller ions. These fragments are analyzed by tandem mass spectrometry (MS/MS), where the resulting spectra are interpreted to identify the sequence of amino acids in the peptide.
The sequencing process works by comparing the observed fragmentation pattern to theoretical models, using algorithms that match the data with potential sequences. This allows researchers to determine the precise order of amino acids, even for previously unknown proteins. One key advantage of this method is its ability to identify novel or modified proteins that may not be present in standard protein databases, making it invaluable for studying organisms with incomplete genetic data.
The Workflow of Peptide and Protein De Novo Sequencing by Mass Spectrometry
The workflow of peptide and protein de novo sequencing by mass spectrometry (MS) is a multi-step process that enables the identification of amino acid sequences without the need for prior sequence knowledge. This approach is highly effective for studying novel proteins or modifications that are not present in existing databases. The process typically follows several key stages:
-
Sample Preparation: The first step in the workflow involves preparing the protein sample. Proteins are extracted from biological samples and then enzymatically digested into smaller peptides, often using enzymes like trypsin. This step is crucial, as it generates the peptide fragments that will later be analyzed by mass spectrometry.
-
Peptide Separation: Once the peptides are generated, they are separated by liquid chromatography (LC) to reduce complexity before being introduced into the mass spectrometer. The LC step ensures that individual peptides are isolated for analysis, increasing the accuracy of the mass spectrometric data.
-
Mass Spectrometry Analysis: In this stage, the separated peptides are ionized and analyzed by the mass spectrometer. The MS system measures the mass-to-charge ratio (m/z) of each ion, providing initial data about the peptide composition. The data generated includes the peptide's molecular weight and charge, which serves as the foundation for subsequent sequencing.
-
Tandem Mass Spectrometry (MS/MS): After initial analysis, the peptides undergo fragmentation in a second stage of mass spectrometry, known as MS/MS. The peptides are broken into smaller fragments, and the mass spectrometer measures the m/z ratios of these fragment ions. The fragmentation patterns are crucial for reconstructing the original peptide sequence.
-
Data Analysis and Sequencing: The final step involves analyzing the MS/MS data to deduce the peptide’s amino acid sequence. Computational algorithms are employed to match the observed fragmentation patterns to known amino acid sequences, and this data is used to infer the sequence of the peptide. De novo sequencing can be particularly useful when no reference sequence is available.
-
Protein Identification: By repeating this process with multiple peptides, researchers can reconstruct the full protein sequence, even identifying post-translational modifications (PTMs). This information is essential for understanding protein function and interactions in biological systems.
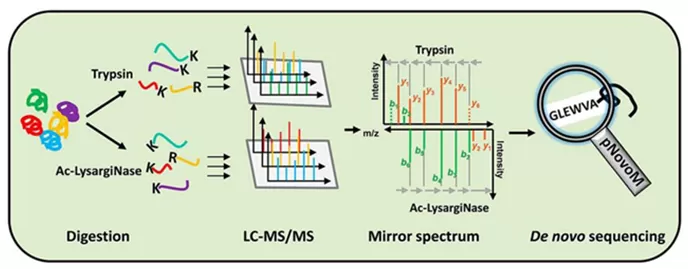
Figure 1. The process of peptide de novo sequencing by mass spectrometry (Hao, et al. 2019).
The Applications of Peptide and Protein De Novo Sequencing by Mass Spectrometry
Peptide and protein de novo sequencing by mass spectrometry (MS) has transformed the landscape of proteomics and is widely applied in various scientific and clinical research areas. This powerful technique allows researchers to determine the amino acid sequences of peptides and proteins, even in the absence of prior genomic or sequence data. The following are some of the key applications where de novo sequencing by mass spectrometry plays a critical role:
-
Identification of Novel Proteins: De novo sequencing is essential for discovering previously unknown proteins, particularly in organisms with incomplete or unsequenced genomes. This is particularly valuable in studying species whose genetic data is not readily available or in exploring novel proteins in areas like microbiology or environmental science. By sequencing peptides directly from the mass spectrum, researchers can identify proteins and gain insights into their functional roles without the need for prior sequence information.
-
Post-Translational Modification (PTM) Analysis: Post-translational modifications (PTMs) such as phosphorylation, glycosylation, and acetylation are crucial for regulating protein function. De novo sequencing by mass spectrometry is an invaluable tool in identifying these modifications, helping scientists understand how PTMs influence cellular processes, protein interactions, and disease mechanisms. This application is particularly important in cancer research, drug development, and neurobiology.
-
Biomarker Discovery for Disease Diagnosis: One of the most promising applications of de novo sequencing is the discovery of novel biomarkers for disease. By analyzing protein samples from patients, scientists can identify unique protein signatures that indicate the presence of specific diseases, such as cancer, neurodegenerative disorders, and metabolic conditions. These biomarkers are vital for developing more accurate diagnostic tools, personalized treatments, and targeted therapies.
-
Drug Discovery and Development: In drug discovery, understanding the structure and function of proteins is key to designing effective therapeutics. De novo sequencing allows researchers to identify and characterize target proteins involved in disease pathways. By elucidating the amino acid sequences of these proteins, scientists can design small molecules or biologics that specifically interact with the target, leading to more efficient drug development.
-
Metabolomics and Systems Biology: De novo sequencing of proteins is often combined with metabolomics and genomics to provide a holistic view of cellular processes. This systems biology approach enables researchers to understand complex biological systems by integrating protein and metabolite data. It helps in identifying biomarkers, uncovering metabolic networks, and studying disease mechanisms at a molecular level, especially in areas like cancer, diabetes, and cardiovascular diseases.
-
Microbial Proteomics: De novo sequencing is crucial in microbial proteomics, where researchers aim to study microorganisms that are difficult to culture or whose genomes have not been sequenced. This technique allows for the identification of microbial proteins directly from environmental or clinical samples, providing insights into microbial diversity, ecological roles, and potential therapeutic targets.
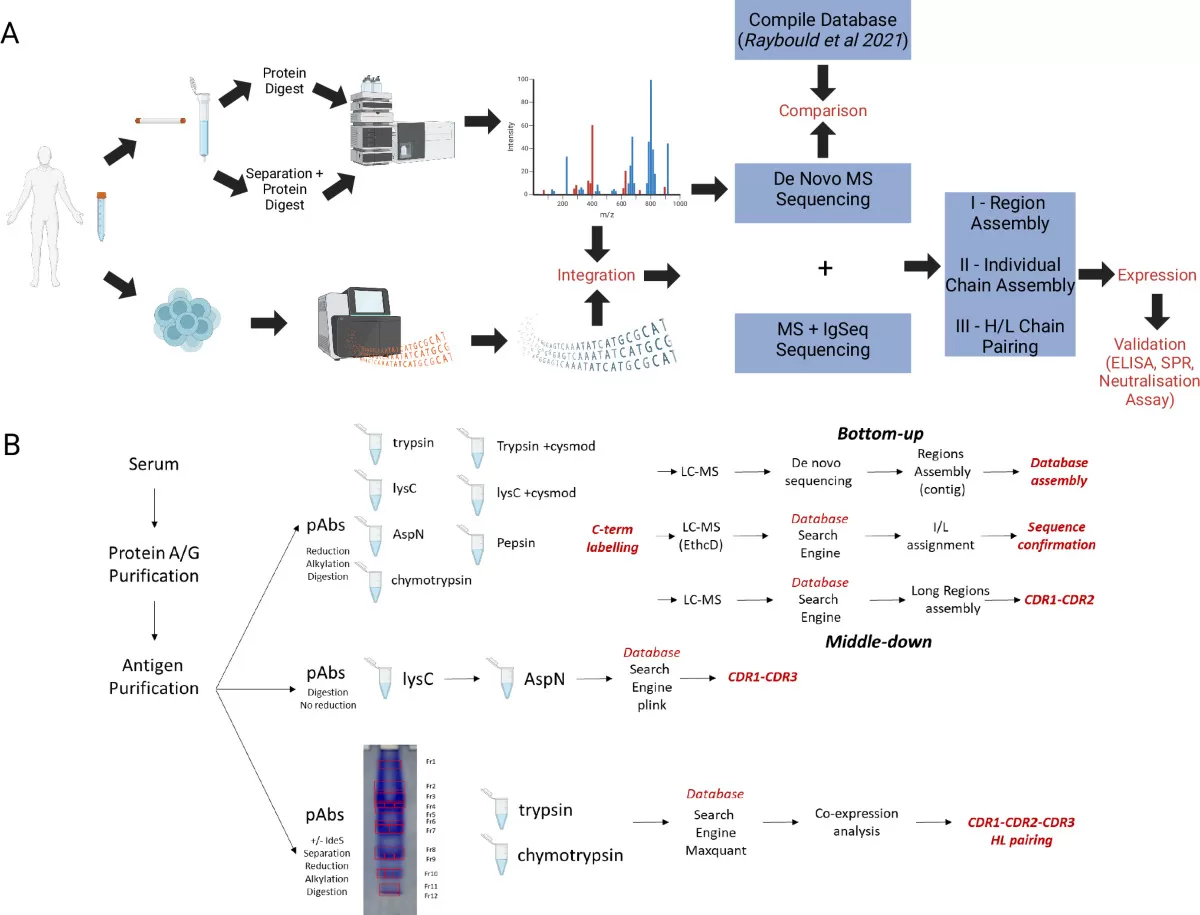
Figure 2. The Applications of Protein De Novo Sequencing by Mass Spectrometry (Le, C. et al. 2024).
References
1. Hao Yang, Yan Chang Li, Ming Zhi Zhao, et al. Precision De Novo Peptide Sequencing Using Mirror Proteases of Ac-LysargiNase and Trypsin for Large-scale Proteomics. Molecular & cellular proteomics: MCP, 2019.
2. Le Bihan, T., Nunez de Villavicencio Diaz, T., Reitzel, C. et al. De novo protein sequencing of antibodies for identification of neutralizing antibodies in human plasma post SARS-CoV-2 vaccination. Nat Commun 15, 8790 (2024).


