Optimal Protein Database Selection: Insights from Experimental Data
To assess the influence of various databases on proteomic data quality, we also searched the proteomic data of serum samples with and without depletion of high-abundance proteins across different databases. Subsequently, the qualitative and quantitative results of protein identification were evaluated.
Serum removing high-abundance proteins
The peptides identified using Swiss-Prot, Proteome, and UniProteinKB were 18,752, 18,527, and 18,936, respectively. Correspondingly, the number of proteins identified were 2,487, 2,661, and 3,769, respectively. Across all three databases, the proportion of proteins and peptides identified was 53.68% and 73.72%, respectively.
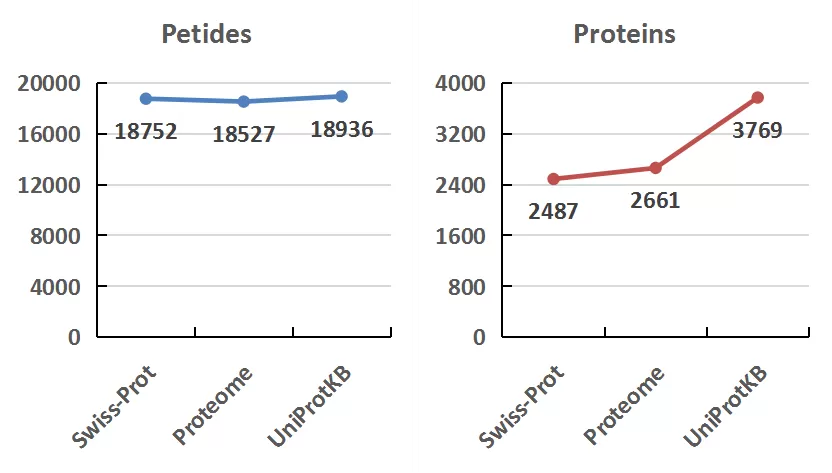
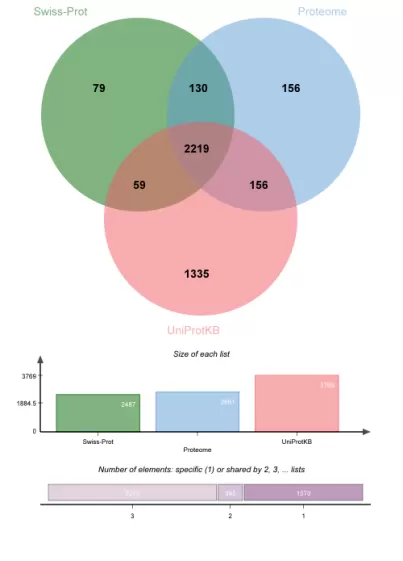
The UniProtKB database identified a notably greater number of proteins compared to Swiss-Prot and Proteome. Among the 1,335 proteins uniquely identified by UniProtKB, 1,172 (87.79%) originated from TrEMBL, 156 from Swiss-Prot, and 7 from Proteome. Notably, 76 were immunoglobulins, and 1,002 (85.49%) proteins lacked gene names.
- Compared to the UniProtKB/Swiss-Prot database, the Proteome and UniProtKB databases respectively yielded a 6.99% and 51.75% increase in identified proteins. While the former showed only a modest increase, the latter exhibited a significant rise.
- Although UniProtKB identified the highest number of proteins, over 85% of them may originate from predicted coding genes, raising doubts about their authenticity.
In summary, the UniProtKB database offers a substantial advantage in detecting proteins in high-abundance proteins removed serum samples, with a 51% increase over Swiss-Prot. Despite concerns about the authenticity of some proteins, UniProtKB still covers over 91.5% of proteins found in Swiss-Prot. Therefore, for high-abundance proteins removed serum samples, it is recommended to prioritize UniProtKB (Swiss-Prot+TrEMBL) for subsequent proteomic data analysis.
Serum with high-abundance proteins
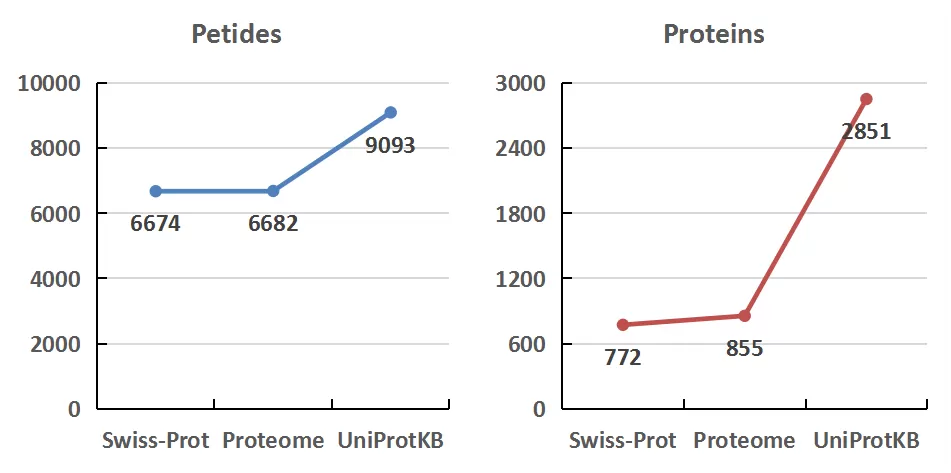
The peptides identified using Swiss-Prot, Proteome, and UniProtKB databases were 6674, 6682, and 9093, respectively. The corresponding identified proteins were 772, 855, and 2815, respectively. The proportion of proteins and peptides identified collectively by all three databases was 19.66% and 53.99%, respectively. The slightly lower number of shared proteins is primarily due to the lower protein identification rates in Swiss-Prot and Proteome compared to UniProtKB.
Analyzing the quantitative missing values across different databases, we observed a consistent trend of missing value variations among all samples. However, the proportion of missing values in the UniProtKB database was higher than that in Swiss-Prot. In the comparison of missing values, Swiss-Prot demonstrated a slight advantage over other databases.
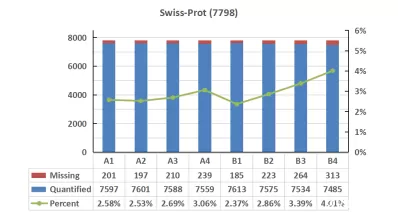
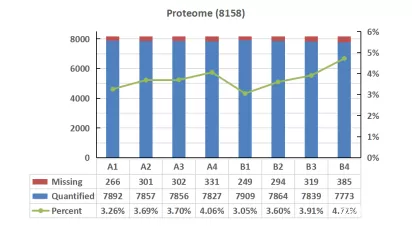
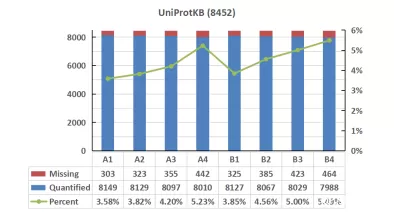
Missing values of proteomics data on serum samples with high-abundance proteins in diffenrent databases
The UniProtKB database identifies a notably higher number of proteins compared to Swiss-Prot and Proteome. Analysis of the 2187 proteins uniquely identified reveals that 2152 (98.40%) proteins originate from TrEMBL, while 29 proteins are sourced from Swiss-Prot and 6 from Proteome. Additionally, there are 103 immunoglobulins, and 1988 (91%) proteins lack gene names.
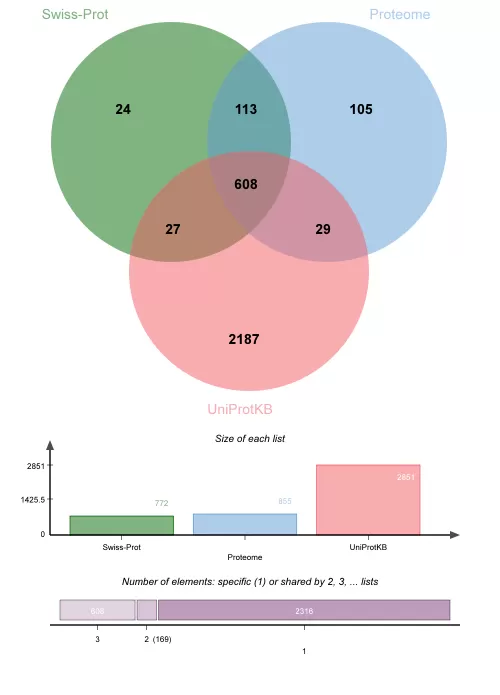
- Compared to the UniProtKB/Swiss-Prot database, the Proteome and UniProtKB databases respectively showed increases of 10.75% and 269.3% in the number of identified proteins. While the former exhibited a modest increase, the latter demonstrated a substantial rise. Despite a higher proportion of missing values in UniProtKB, post-removal of proteins with missing values, 2016 proteins were retained, significantly surpassing those identified by Swiss-Prot and Proteome.
- UniProtKB exhibits a notable advantage in detecting proteins in blood samples with high-abundance proteins, with a 269.3% increase over Swiss-Prot. Despite potential authenticity concerns, UniProtKB still covers over 82% of Swiss-Prot proteins. Moreover, despite a higher proportion of missing values in UniProtKB, the remaining protein count after removal is still substantial.
In conclusion, while UniProtKB may contain proteins not manually verified, it largely encompasses Swiss-Prot information and provides additional data. Thus, for blood samples with high-abundance proteins, prioritizing UniProtKB (Swiss-Prot+TrEMBL) is advisable for subsequent proteomic analysis.
Conclusion: Best Practices for Protein Database Selection
- The UniProtKB database is larger than Swiss-Prot, with the additional proteins observed in empirical data mostly originating from predicted protein translations of coding genes. These proteins are typically generated from a single gene through various biological events (such as alternative promoters, alternative splicing, alternative translation start sites, ribosomal frameshifting, etc.), with no direct evidence of protein existence.
- For tissue/cellular samples, the differences in detected proteins among Swiss-Prot, Proteome, and UniProtKB databases are minor. Considering the higher reliability of protein information in the Swiss-Prot database, it is recommended to use the Swiss-Prot database for subsequent protein identification analysis in human cells/tissues.
- For plasma/serum samples, the detection of proteins in the UniProtKB database is significantly increased compared to other databases. Although the authenticity of some proteins may be questionable, UniProtKB covers the vast majority of information available in Swiss-Prot. Therefore, it is advisable to use the UniProtKB database for subsequent protein identification analysis in human serum/plasma samples.


