Omics Insights Unveiling PCSK9's Role in Heart Transplant Rejection
Introduction: The Importance of Heart Transplantation and the Challenge of Rejection
Heart transplantation is the most effective treatment for patients with refractory end-stage heart disease. However, heart transplant rejection is a major complication that limits long-term graft survival, characterized by massive infiltration of proinflammatory macrophages and effector T cells, along with cardiomyocyte damage. Previous studies have primarily focused on immune cells, leaving the role of nonimmune organs and related molecules in heart transplant rejection largely undefined.
PCSK9: A Potential Game Changer in Transplant Medicine
PCSK9 (proprotein convertase subtilisin/kexin type 9) is the ninth member of the proprotein convertase family. Mainly expressed in the liver, it has been identified as a significant factor in hypercholesterolemia. Due to its functional characteristics, PCSK9 has been targeted for lowering cholesterol levels and reducing cardiovascular events. Recent studies also have shown that PCSK9 inhibitors are safe and effective in lowering cholesterol in patients with hypercholesterolemia after heart transplantation. Additionally, an increasing number of functions of PCSK9 have been found, such as promoting atherosclerosis, aggravating sepsis and viral infection, and regulating tumor immunity. Moreover, serum PCSK9 levels have been observed to increase in patients post-transplant. However, the role of PCSK9 in rejection and the effect of PCSK9 ablation on immune cells after transplantation remain unclear.
Multi-Omics Approach: Decoding the Mechanisms of Rejection
A research paper published in Circulation Immune Regulation of the Liver Through the PCSK9/CD36 Pathway During Heart Transplant Rejection (article resource) reveals a novel mechanism for immune regulation by the liver through the PCSK9/CD36 pathway during heart transplant rejection (HTR). This pathway influences the phenotype and function of macrophages, suggesting that its modulation could be a potential therapeutic target to prevent HTR. The study combined multiorgan histological and transcriptome analyses with multiomics and single-cell RNA sequencing of the liver during HTR. Metwarebio, a leading CRO in metabolomics and multi-omics detection services, provided the transcriptome sequencing, proteomics, and metabolomics detection for this research. Let’s delve into this article to see how this outstanding research was conducted.
PCSK9 Ablation Attenuates HTR in Mice
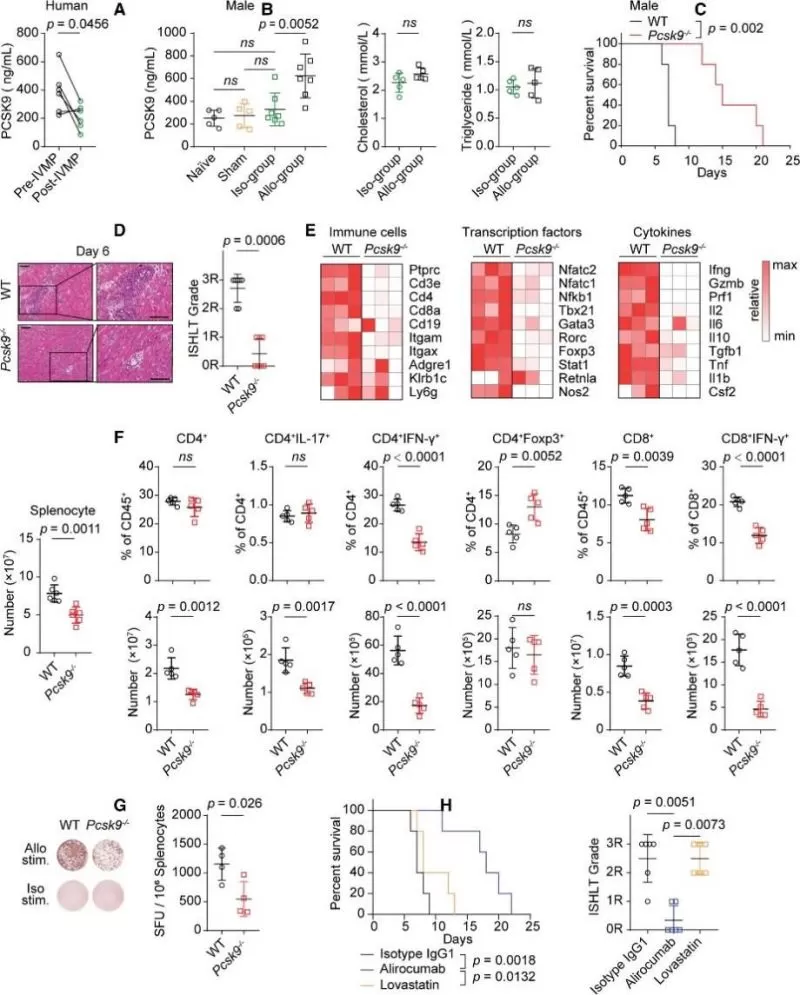
To investigate the role of PCSK9 during HTR, the authors measured the expression of PCSK9 in serum obtained from 6 patients who had heart transplant with rejection and the same recipients after intravenous methylprednisolone treatment. The results showed that serum PCSK9 levels were significantly reduced after intravenous methylprednisolone treatment compared with before intravenous methylprednisolone treatment (Figure 1A). Meanwhile, heart transplantation was performed in mice with full MHC (major histocompatibility complex) mismatch, revealing that serum PCSK9 levels increased in the allogroup mice on day 6 post-transplant compared to naive, sham-operated, and isogroup mice (Figure 1B). Blood lipid levels showed no differences between the isogroup and allogroup (Figure 1B). Next, using Pcsk9 knockout mice in murine heart transplantation showed that Pcsk9 knockout prolonged cardiac allograft survival in both male and female mice and reduced the infiltration of inflammatory cells and myocyte damage (Figure 1C and 1D). RNA-seq analysis revealed that the levels of genes encoding immune cells, transcription factors, and cytokines associated with inflammation were significantly decreased in grafts from Pcsk9–/– mice (Figure 1E). Pcsk9 knockout alleviated splenomegaly and inhibited the expansion of splenocytes during the alloimmune response (Figure 1F). On day 6 post-transplant, flow cytometry showed a decrease in the percentages of IFN-γ–producing CD4+ and CD8+ effector T cells, but an increase in the percentage of splenic regulatory T cells in Pcsk9–/– mice (Figure 1F). Pcsk9 knockout also induced a decrease in the percentage of splenic IFN-γ–producing donor-reactive cells, as detected by an enzyme-linked immunospot assay (Figure 1G). Similarly, neutralizing PCSK9 with alirocumab in mice had comparable effects on graft survival and the alloimmune response in the graft and spleen (Figure 1H). Together, these data indicated that PCSK9 participated in the alloimmune response after heart transplantation.
Histological and Multiomics Analyses of Multiple Organs in Recipients during HTR
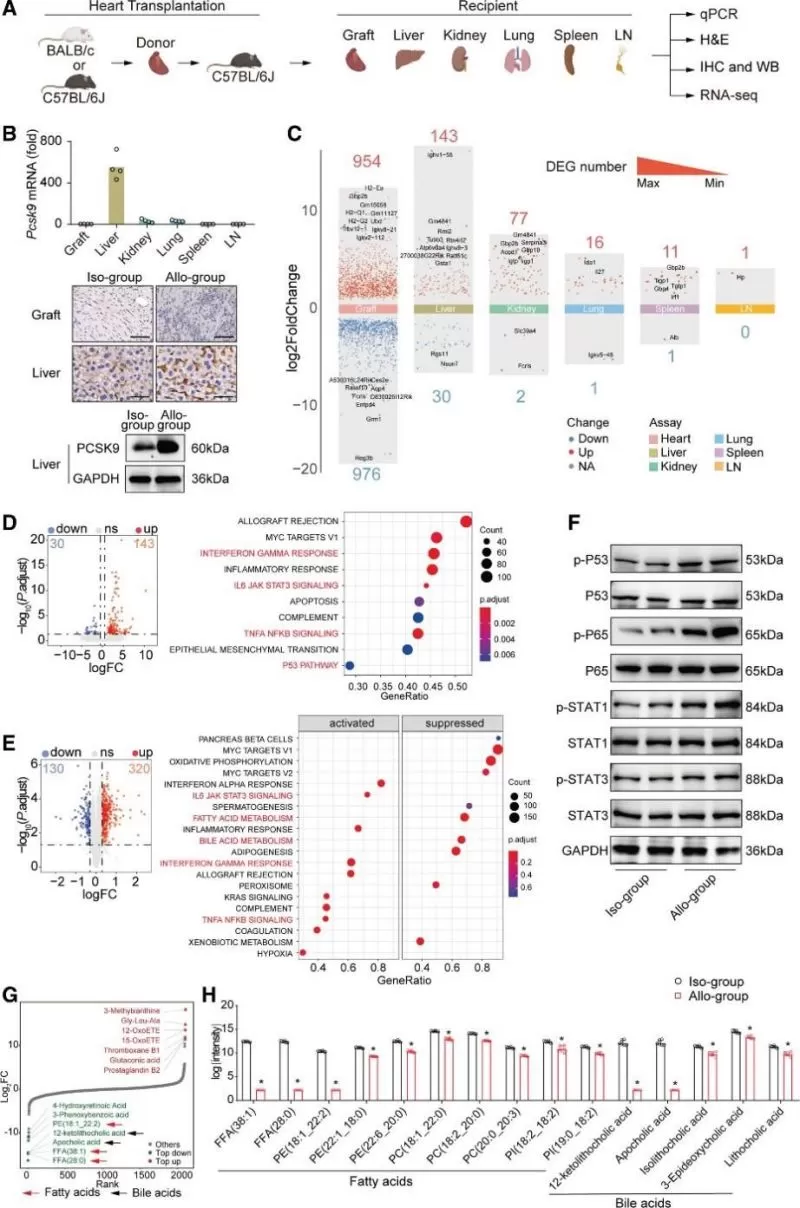
To explore the origin of PCSK9 after transplantation, the histological characteristics and Pcsk9 gene and protein expression in multiple organs (graft, liver, kidney, lung, spleen, lymph node) from recipient mice were analyzed (Figure 2A). Through quantitative polymerase chain reaction screening, a significant increase in Pcsk9 mRNA expression was observed in the liver of allogroup recipient mice compared to other organs (Figure 2B). Immunohistochemistry assays confirmed that PCSK9 was primarily expressed in the recipient liver after heart transplantation and was significantly upregulated in the recipient liver in the allogroup compared to the isogroup (Figure 2B). Western blotting also verified the upregulation of PCSK9 in livers during HTR (Figure 2B). Furthermore, multiorgan RNA-seq analysis revealed a higher number of differentially expressed genes (DEGs) in the liver than in other recipient organs (Figure 2C and Figure S8B). Using a multiomics approach (transcriptomics, proteomics, and metabolomics) to study the liver during HTR, the results suggested that fatty acid and bile acid metabolism in the liver was inhibited during HTR (Figure 2, D-H). This demonstrated the upregulation of PCSK9 expression and highlighted differences in inflammatory and metabolic pathways in recipient livers during HTR.
Single-Cell RNA-Seq Analysis of the Liver in Mice during HTR
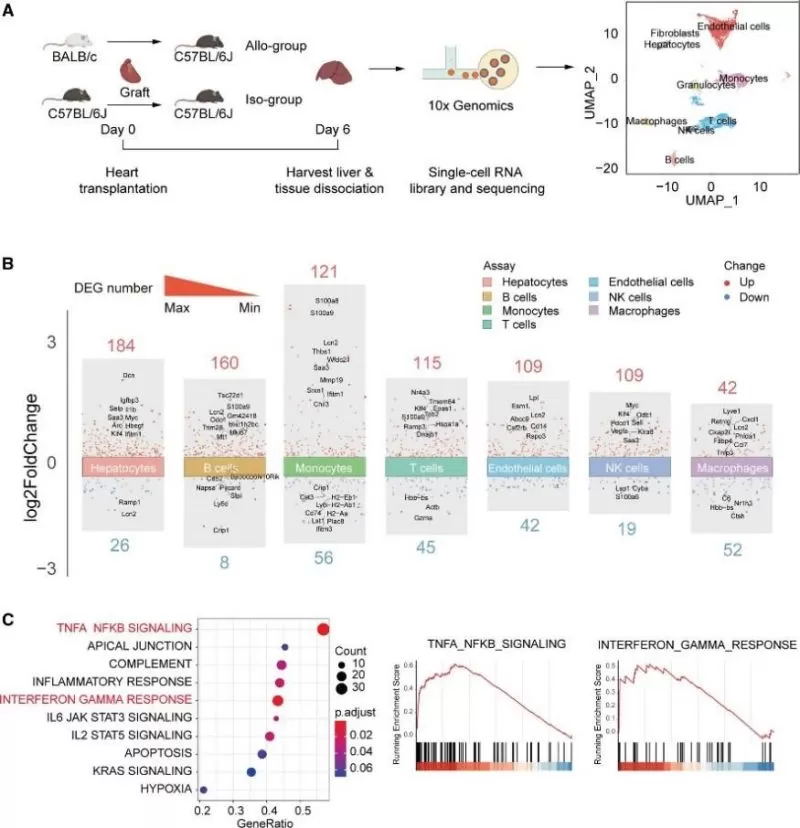
The liver is a multifunctional organ composed of various cells, including hepatocytes, Kupffer cells, stellate cells, and endothelial cells. Single-cell RNA-seq analysis was performed on recipient livers during HTR. The cells were clustered into 9 groups based on the expression of canonical markers (Figure 3A). The clusters, generated based on cell type and corresponding nomenclature, were visualized using uniform manifold approximation and projection (Figure 3A). As shown in Figure 3B, the number of DEGs in hepatocytes was much larger than in other cells. There were 184 significantly upregulated genes and 26 downregulated genes in hepatocytes of the recipient liver between the allogroup and isogroup. DEGs were also enriched in pathways including the TNFA_NFKB_SIGNALING, INTERFERON_GAMMA_RESPONSE, FATTY_ACID_METABOLISM, and BILE_ACID_METABOLISM pathways (Figure 3C), which is consistent with the results of the multiomics analysis of the liver (Figure 2).
TNF-α and IFN-γ Synergistically Increase PCSK9 Expression in Hepatocytes through SREBP2
To elucidate whether activation of inflammatory pathways in hepatocytes caused by the cytokine storm during HTR promotes PCSK9 expression, a multiplex cytokine assay of serum from recipient mice was performed on day 6 post-transplant. The levels of proinflammatory cytokines (e.g., IFN-γ, TNF-α, GM-CSF, VCAM-1, and M-CSF) were found to be significantly increased (Figure 4A). Next, primary mouse hepatocytes were stimulated with these cytokines in vitro, revealing that TNF-α and IFN-γ, but not GM-CSF, VCAM-1, or M-CSF, induced the mRNA expression of PCSK9 (Figure 4B). Additionally, ELISA showed that PCSK9 levels significantly increased in the culture supernatant of hepatocytes after 24 hours of stimulation with TNF-α and the combination of TNF-α and IFN-γ (Figure 4C).
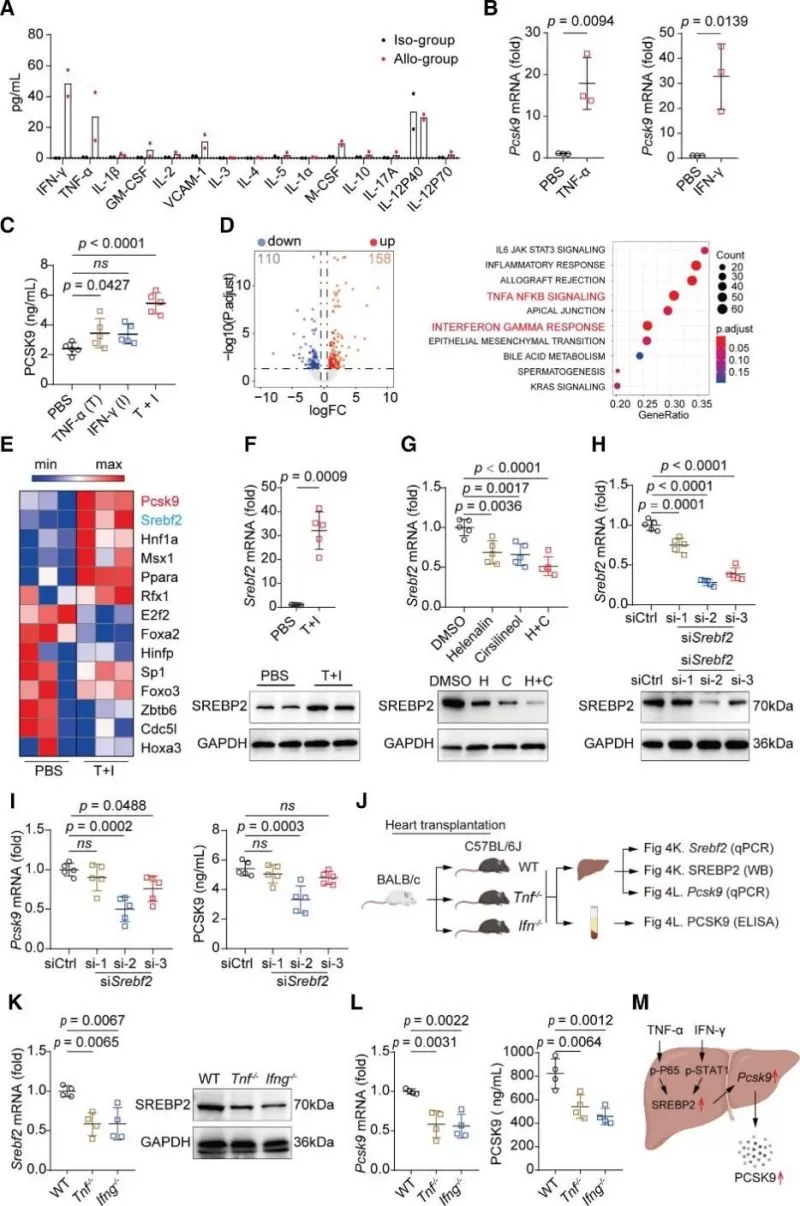
To understand how TNF-α and IFN-γ together induce PCSK9 expression, RNA-seq was performed on hepatocytes with or without combined treatment, identifying 268 DEGs, including 158 upregulated and 110 downregulated genes (Figure 4D). Among the transcription factors previously reported to promote PCSK9 expression, Srebf2 was significantly upregulated in hepatocytes after the combined treatment (Figure 4E). It was verified that an SREBP2-independent regulatory mechanism controls LDLR expression in TNF-α and IFN-γ stimulated hepatocytes (Figure 4, F-I). Further in vivo studies (Figure 4J)showed that Tnf or Ifn deficiency not only inhibited the expression of SREBP2 in the recipient livers but also reduced liver Pcsk9 mRNA expression and the serum PCSK9 level during HTR (Figure 4K and 4L). Together, these data suggest that TNF-α and IFN-γ synergistically promote PCSK9 expression in hepatocytes through SREBP2 (Figure 4M).
PCSK9 Influences CD36 Expression and Macrophage Function
To elucidate the mechanism underlying the proinflammatory effect of PCSK9 during HTR, previously reported targets of PCSK9 were induced in cardiac allografts during rejection by analyzing a microarray dataset containing data from human endomyocardial biopsies (GSE124897). Among the investigated targets, only CD36 expression was significantly different in endomyocardial biopsies between rejection and no-rejection grafts (Figure 6A). To further investigate the localization of CD36 at the cellular level in cardiac allografts, single-cell RNA-seq analysis of CD45+ graft-infiltrating immune cells were performed and the results showed that Cd36 was mainly expressed in graft-infiltrating macrophages (Figure 6B). Next, the expression of CD36 on macrophages in cardiac allografts was verified by using immunofluorescence staining (Figure 6C). Further in vivo studies suggested that PCSK9 not only controls CD36 expression and fatty acid uptake but also enhances the proinflammatory phenotype and function of macrophages (Figure 6, D-G).
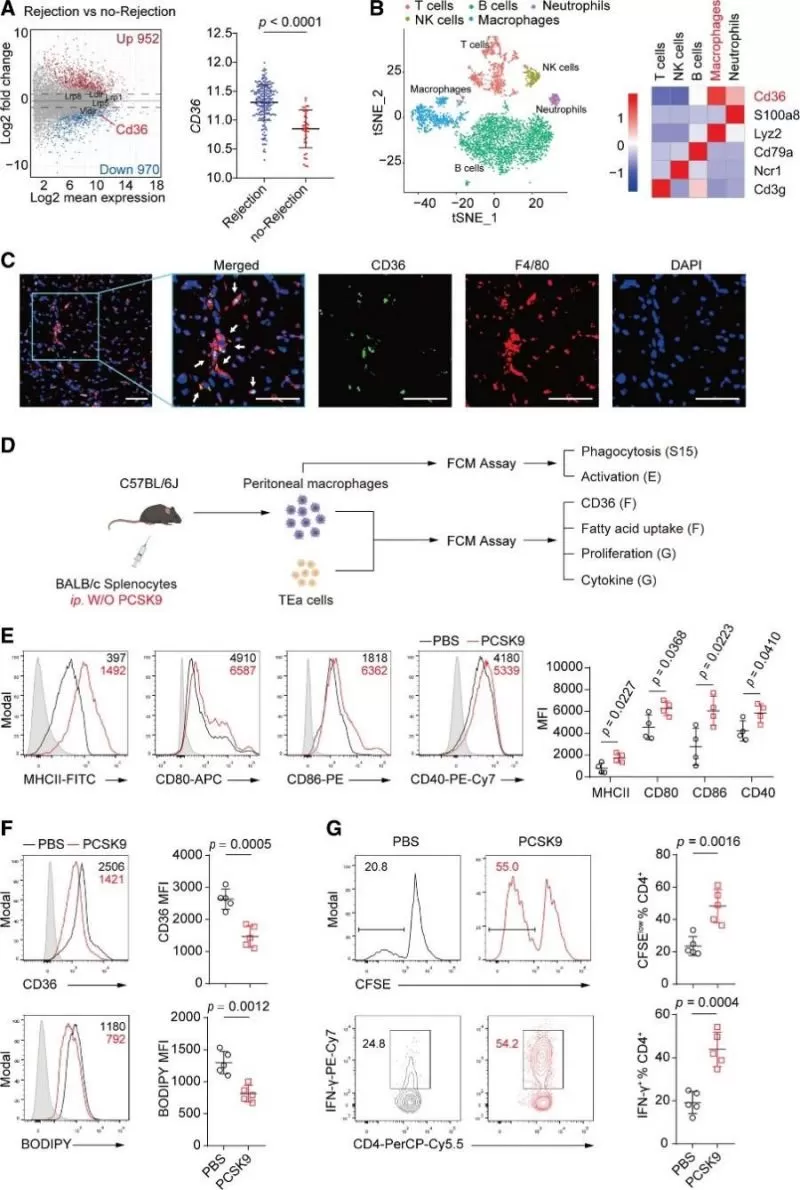
Conclusion: Advancing Transplant Medicine with Omics Research
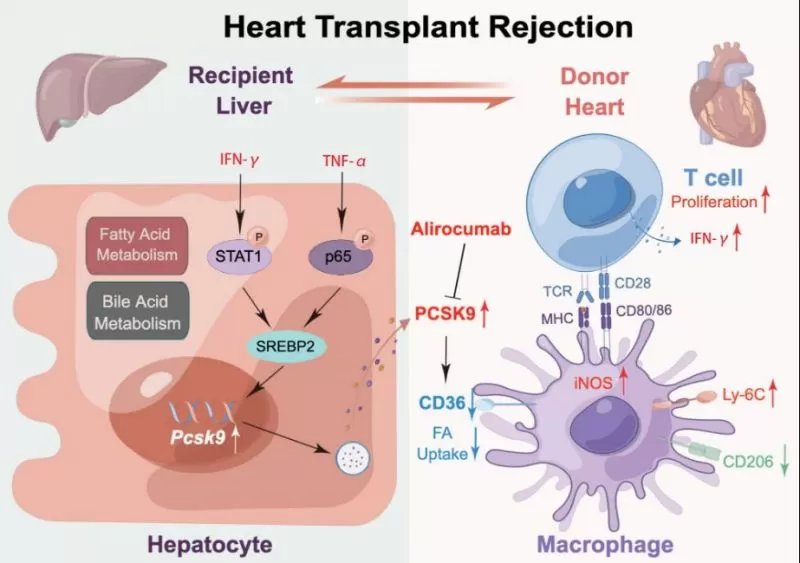
This study found that during HTR, TNF-α and IFN-γ synergistically promoted PCSK9 expression in hepatocytes through the transcription factor SREBP2, resulting in high serum PCSK9 levels. Then, the increased PCSK9 inhibited CD36 expression and fatty acid uptake to favor proinflammatory macrophages, which promoted the proliferation and IFN-γ production of donor-reactive T cells (Figure 8). This study reveals a novel mechanism for immune regulation by the liver during HTR through the PCSK9/CD36 pathway, which influences the phenotype and function of macrophages, and the study also suggests that the modulation of this pathway may be a potential therapeutic target in HTR.
Enhance Your Research with MetwareBio's Cutting-Edge Multi-Omics Services
The study's comprehensive approach, integrating multiorgan histological and transcriptome analyses with multiomics and single-cell RNA sequencing, highlights the critical role of multiomics services in uncovering complex biological pathways and mechanisms. The utilization of these advanced techniques provided a detailed and holistic understanding of the molecular interactions at play, underscoring the value of multiomics in driving forward innovative and impactful biomedical research. MetwareBio is a multiomics CRO focusing on developing and applying innovative multiomics technologies to life science and health research. With a dedicated commitment to data quality and a nuanced understanding of the unique nature of each project, MetwareBio offers tailored metabolomics, proteomics and multi-omics combination analyses services to suit diverse needs. Whether it's small-scale endeavors or large population studies, our workflows are adept at accommodating varying sample sizes and project scopes. Our extensive experience, reflected in over 4000 completed projects, underscores our proficiency in delivering reliable results. At MetwareBio, we prioritize collaboration, guiding researchers from sample extraction to data analysis to ensure their research goals are met with precision and efficiency. Please don't hesitate to reach out if you have any requirements or inquiries!
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


