Olink Proteomics Explained: Principles, Advantages, and Comparison with MS-Based Methods
Proteins, as the primary executors of cellular functions, provide critical insights into the underlying mechanisms of diseases, biological processes, and cellular responses. Proteomics is the large-scale study of proteins, particularly their functions, structures, and interactions within biological systems. In the rapidly evolving field of proteomics, the quest for advanced, high-resolution, and high-throughput technologies continues to drive innovation. Among these, Olink proteomics technology represents a significant advancement in protein analysis. This blog aims to provide a comprehensive introduction to Olink proteomics technology, outlining its principles and comparing its advantages and limitations relative to other proteomics methods, such as mass spectrometry (MS)-based techniques. By emphasizing the distinctive features of Olink technology, we seek to guide researchers in selecting the most appropriate method for their specific research goals. Whether investigating novel biomarkers or enhancing clinical diagnostics, a clear understanding of these differences will aid in choosing the optimal proteomics approach for achieving accurate and impactful results.
Introduction to Olink Proteomics
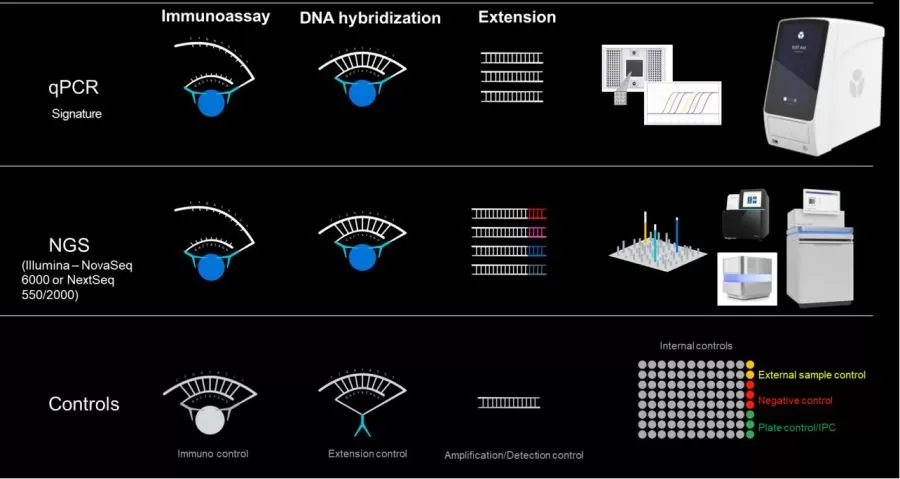
 Olink proteomics technology is a protein detection method based on Proximity Extension Assay (PEA) technology. PEA successfully combines antibody-based immunoassays with the powerful features of PCR and quantitative real-time PCR (qPCR) or next-generation sequencing (NGS) platforms, creating a multiplexed and highly specific approach capable of simultaneously quantifying up to 92–5400+ protein biomarkers.
Olink proteomics technology is a protein detection method based on Proximity Extension Assay (PEA) technology. PEA successfully combines antibody-based immunoassays with the powerful features of PCR and quantitative real-time PCR (qPCR) or next-generation sequencing (NGS) platforms, creating a multiplexed and highly specific approach capable of simultaneously quantifying up to 92–5400+ protein biomarkers.
Principles of PEA Technology
The core of PEA is a dual-recognition immunoassay in which two matched antibodies bind simultaneously to a target and are uniquely labeled with DNA oligonucleotides. When the target protein is present, the antibodies are brought into proximity, allowing their DNA oligonucleotides to hybridize, serving as templates for a DNA polymerase-dependent extension step. This creates a double-stranded DNA “barcode” that is unique to the specific antigen and quantitatively proportional to the initial concentration of the target protein. Following hybridization and extension, PCR amplification is performed immediately, and the amplicons are finally quantified using microfluidic qPCR.
Olink's patented PEA technology employs a pair of specifically matched antibodies for each antigen (protein), ensuring the specificity of immunoreactions. Furthermore, PEA technology enhances specificity by attaching unique oligonucleotide pairs to each antibody pair, which, through DNA pairing specificity, prevents cross-reactivity at the immunological level during multiplex detection. Ultimately, only antibody pairs that specifically bind to their corresponding antigens form a double-stranded DNA molecule, which is then extended into a PCR-amplifiable nucleic acid double strand and detected by downstream qPCR or NGS.
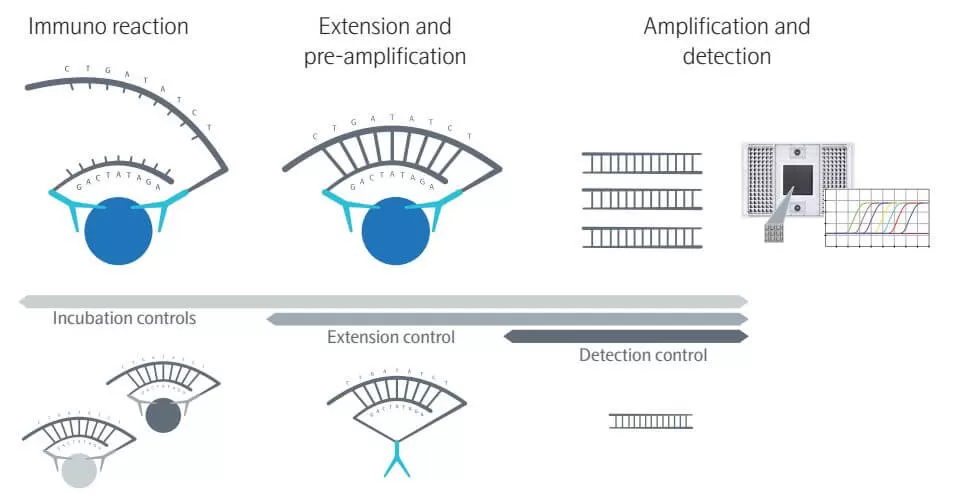
Utilizing a high-throughput microfluidic qPCR platform, PEA technology can simultaneously detect 96 proteins (92 target proteins and 4 internal controls) in a single sample well using just 1 µL of fluid. Using an NGS platform, PEA technology can detect up to 5400+ proteins in a single sample well using 8 µL of fluid, enabling highly multiplexed, high-sensitivity protein detection.
Advantages of Olink Proteomics
1. High Specificity
Leveraging patented PEA technology, robust quality control design, and thorough validation, all data exhibit exceptional specificity. Each target protein is a well-defined biomarker, overcoming the specificity challenges of traditional methods.
2. High Throughput
Capable of rapidly detecting 45 to 5400+ protein biomarkers across multiple samples.
3. Low Sample Volume
Requires less than 8 µL of sample per assay.
4. High Sensitivity
Detection sensitivity reaches fg/mL levels, allowing the identification of hundreds to thousands of low-abundance, disease-related proteins at the omics level.
5. Wide Dynamic Range
Spanning 10 logs, the dynamic range covers high, medium, and low-abundance proteins, with a particular strength in detecting low-abundance proteins.
6. Automation
Automated sample processing offers exceptional ease, accuracy, and reproducibility.
7. High Reproducibility
Utilizing mature qPCR and NGS technologies, this approach provides excellent reproducibility and high data quality, meeting the demands of big data analysis and clinical translational applications.
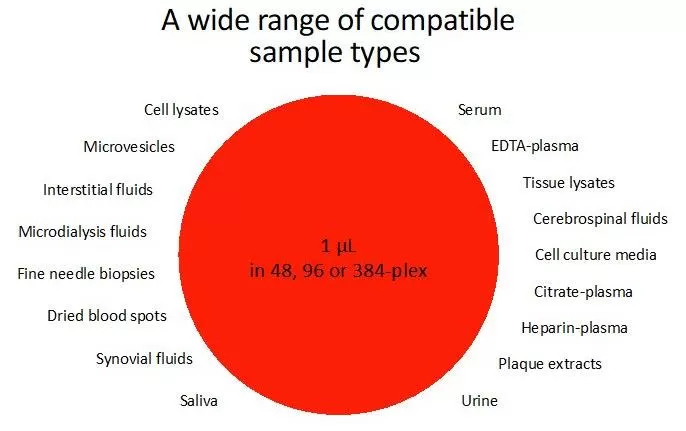
8. Compatibility with Diverse Samples
Suitable for a broad range of sample types, including serum, various plasmas, tissue lysates, cell culture media, cerebrospinal fluid, plaque lysates, urine, cell lysates, microvesicles/exosomes, interstitial fluid, microdialysis fluid, fine-needle biopsy samples, dried blood spots, synovial fluid, saliva, aqueous humor, and more. Its outstanding sensitivity is particularly advantageous for fluid samples.
Olink Proteomics vs. MS-Based Proteomics
In the realm of proteomics, both Olink proteomics and mass spectrometry (MS)-based proteomics are powerful techniques, each offering distinct advantages and addressing different research needs. Here, we compare these two approaches to help researchers make informed decisions based on their specific objectives.
1. Methodology
- Olink Proteomics: Olink utilizes Proximity Extension Assay (PEA), which involves two specific antibodies labeled with unique DNA oligonucleotides. When the antibodies bind to the target protein, the DNA oligonucleotides come into close proximity, allowing them to hybridize and serve as a template for DNA polymerase-dependent extension. This generates a unique DNA barcode that is amplified and quantified using PCR or next-generation sequencing (NGS). This approach is highly specific and allows simultaneous measurement of a large number of proteins.
- MS-Based Proteomics: Mass spectrometry-based proteomics relies on the ionization of proteins and their fragments, followed by separation based on mass-to-charge ratio. The resulting mass spectra provide detailed information on protein identity and abundance. Techniques such as liquid chromatography-mass spectrometry (LC-MS) are commonly used to separate proteins before MS analysis, providing comprehensive insights into complex protein mixtures.
2. Specificity and Sensitivity
- Olink Proteomics: Olink’s PEA technology excels in specificity due to its use of matched antibody pairs, minimizing cross-reactivity and providing accurate quantification of target proteins. It is highly sensitive, capable of detecting proteins at femtogram per milliliter (fg/mL) levels, which is particularly useful for identifying low-abundance biomarkers.
- MS-Based Proteomics: While MS-based proteomics offers high sensitivity and can detect a broad range of proteins, its specificity can be influenced by sample complexity and ionization efficiency. Some proteins may be difficult to detect due to interference or low ionization efficiency, which can affect the overall accuracy of quantification.
3. Throughput and Coverage
- Olink Proteomics: Olink technology supports high-throughput analysis, enabling the simultaneous detection of up to 5400+ proteins from a single sample. This multiplexing capability is ideal for large-scale studies requiring the measurement of numerous biomarkers in parallel.
- MS-Based Proteomics: MS-based methods, especially those using advanced techniques like Data-Independent Acquisition (DIA) on Bruker TIMS TOF series of mass spectrometers, can achieve even greater throughput. DIA-MS can detect and quantify over 10,000 proteins in a single run, providing deeper proteomic coverage than Olink. This makes MS-based proteomics highly suitable for comprehensive proteome profiling. However, the complexity of data analysis in MS-based proteomics, particularly when dealing with such large datasets, can be a limiting factor compared to Olink’s more streamlined analysis.
4. Sample Volume and Preparation
- Olink Proteomics: Olink’s approach requires a minimal sample volume (as low as 1 µL), which is advantageous for studies with limited or precious samples. The sample preparation involves adding specific reagents and performing a straightforward extraction process. Although Olink proteomics is versatile, Olink is primarily optimized for liquid samples, certain sample types or conditions may still pose challenges. For instance, very complex biological matrices or highly variable samples might affect the performance or accuracy of the assay.
- MS-Based Proteomics: While MS-based proteomics typically requires larger sample volumes compared to Olink, it excels at handling complex biological matrices, including tissues, cells, and fat. The sample preparation process, although more intricate—often involving protein extraction, digestion, and cleanup—is highly efficient and well-suited for a wide range of sample types. This capability allows MS-based proteomics to deliver reliable results even with challenging sample types.
5. Applications and Use Cases
- Olink Proteomics: Olink is well-suited for targeted biomarker discovery, clinical diagnostics, and research where high specificity and multiplexing are critical. It is particularly effective in scenarios where precise quantification of specific proteins is required.
- MS-Based Proteomics: MS-based proteomics is advantageous for comprehensive proteome characterization, discovery-based studies, and detailed protein profiling. It is often used for exploratory research where broad and in-depth analysis of the proteome is necessary.
MetwareBio’s Proteomics Services
MetwareBio is a multiomics CRO focusing on developing and applying innovative multiomics technologies to life science and health research. With a dedicated commitment to data quality and a nuanced understanding of the unique nature of each project, MetwareBio offers tailored single-omics and multi-omics combination analyses services to suit diverse needs. Whether it's small-scale endeavors or large population studies, our workflows are adept at accommodating varying sample sizes and project scopes. MetwareBio is offering the following comprehensive MS-based proteomics services:
- DIA Quantitative Proteomics: Offers high-throughput, highly accurate protein quantification, making it ideal for large-scale comprehensive proteome profiling.
- DDA Quantitative Proteomics: Delivers high-sensitivity and precise protein identification, making it well-suited for exploratory studies aimed at identifying the maximum number of unique proteins from complex biological samples.
- Serum/Plasma Quantitative Proteomics: Focuses on the analysis of proteins in blood samples, offering valuable insights into disease biomarkers, immune responses, and physiological states due to its minimally invasive nature and accessibility.
- Phosphoproteomics: Enables quantitative analysis of protein phosphorylation modifications, crucial for understanding cellular signaling pathways and post-translational modifications, playing a key role in cancer research and drug development.
If you are interested in proteomics research, please don't hesitate to reach out.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


