Multi-Omics Association Analysis (III): Association Analysis of Proteomics and Transcriptomics
The Role of Proteomics and Transcriptomics in Gene Expression
In living organisms, the primary processes of gene expression involve transcription and protein synthesis, with mRNA serving as the intermediary and proteins acting as the executors of gene functions. To fully understand the regulatory mechanisms governing gene expression, it is crucial to conduct association analyses between mRNA and proteins.
Approach for Combined Analysis of Proteomics and Transcriptomics
The integration of proteomics and transcriptomics is a multi-omics research approach that combines and compares mRNA and protein data from the same biological samples, thereby elucidating the regulatory mechanisms at both transcriptional and translational levels. This analysis encompasses not only mRNA expression patterns but also the abundance and modification states of proteins, offering a more comprehensive perspective on gene expression regulation. Given that post-transcriptional and post-translational regulations can lead to discrepancies between mRNA and protein levels, combined analyses provide a more accurate reflection of the dynamic changes and biological functions involved in gene expression. Moreover, this approach is essential for understanding complex biological processes, clarifying disease mechanisms, and informing medical diagnostics and drug development. With advancements in technologies such as mass spectrometry, proteomics has become an indispensable tool for investigating gene functions and disease mechanisms, and its integration with transcriptomics is emerging as a prominent research trend.
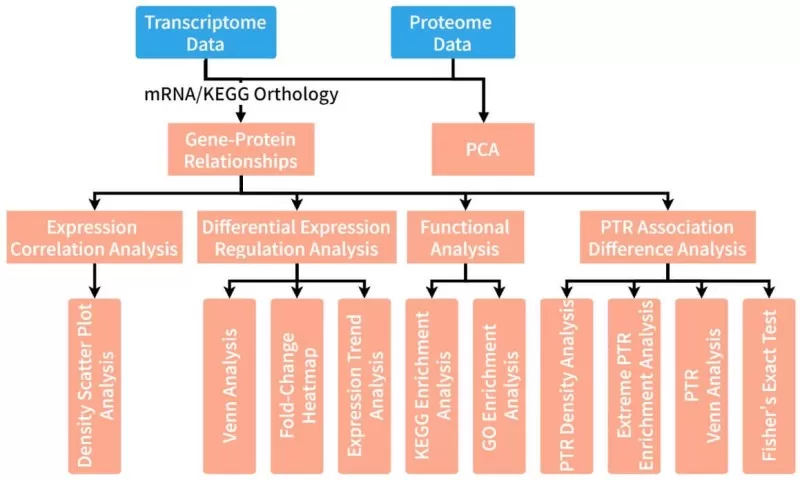
Display of Selected Analysis Results
1) Expression Correlation Analysis: mRNA and Protein Dynamics
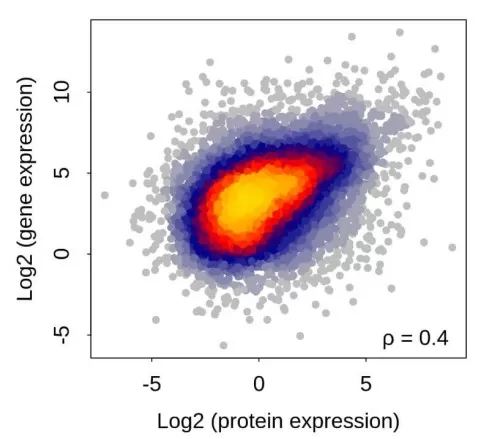
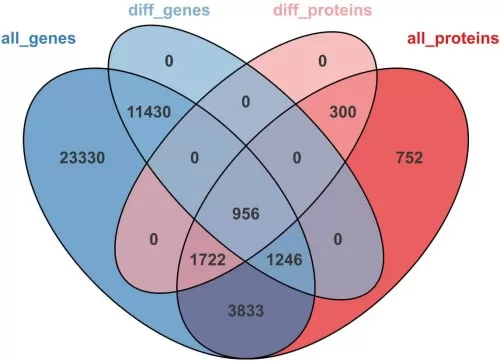
2) Differential Expression Association Analysis: Methods and Visualization
These analyses mainly include Venn diagram, clustering heatmap, volcano plot (refer to the volcano plot from the proteomics and modification association analysis), and nine-quadrant plot (refer to the nine-quadrant plot from the proteomics and metabolomics association analysis).
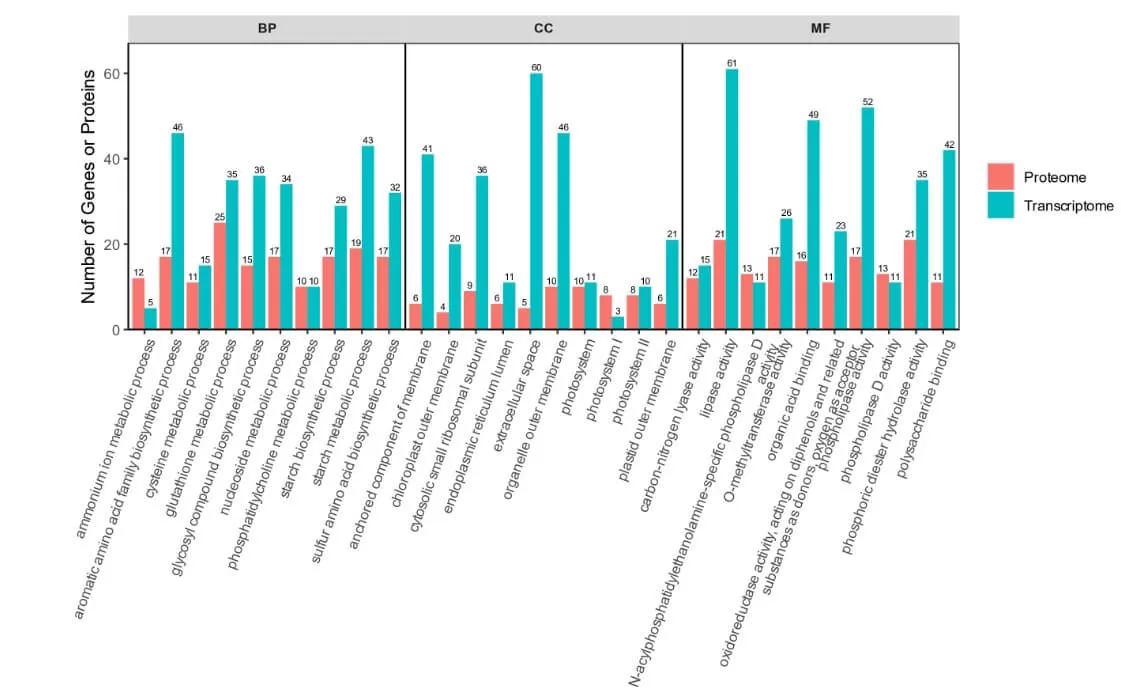
3) Differential GO/KEGG Enrichment Analysis: Insights into Biological Processes
The enrichment bubble plot is similar to the functional comparison bubble plot from the proteomics and modification association analysis.
4) PTR Analysis: Revealing Protein Translation Efficiency
The Protein-to-mRNA Ratio (PTR) is an indicator of gene translation efficiency, calculated by dividing the protein expression level (represented as log2 LFQ values) by its mRNA transcription level (represented as log2 FPKM values). Analyzing PTR values aids in revealing the cellular regulatory mechanisms influencing protein synthesis rates.
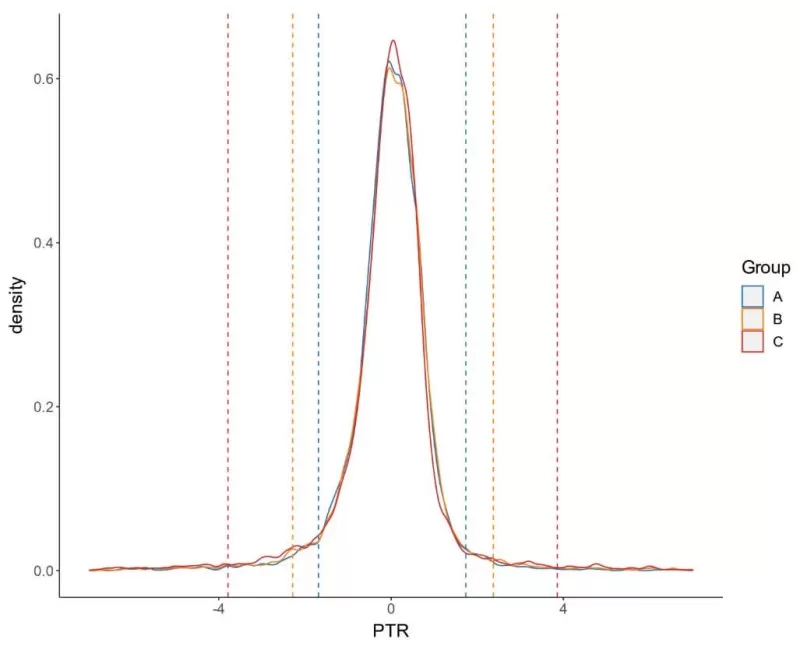
i) PTR Density Distribution
A high PTR value typically indicates effective translation processes and a high protein synthesis rate, while a low PTR value may suggest regulatory control over translation or reduced protein synthesis efficiency.
ii) Differential Proteins (Genes) and Extreme PTR Fisher's Exact Test
Fisher’s exact test is a method suitable for analyzing the statistical significance of contingency tables, used to determine whether a significant association exists between two categorical variables. This method assesses the association between differential proteins or genes and extreme PTR values.

Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


