MALDI, DESI, or SIMS? How to Choose the Best MSI Techniques for Spatial Metabolomics
Spatial metabolomics is transforming our ability to study the distribution of metabolites within biological tissues. By combining the analytical power of metabolomics with advanced imaging techniques, researchers can uncover intricate chemical landscapes and gain critical insights into biology, medicine, and agriculture.
Selecting the right mass spectrometry imaging (MSI) technology is a crucial decision for researchers in spatial metabolomics. Each technique—MALDI (Matrix-Assisted Laser Desorption/Ionization), DESI (Desorption Electrospray Ionization), and SIMS (Secondary Ion Mass Spectrometry)—offers unique advantages, making them suitable for different applications. But how do you determine which is the best fit for your specific research needs? For a quick comparison of MSI Techniques, refer to the comparison of MALDI, DESI, and SIMS in Spatial Metabolomics table below.
In this guide, we’ll explore the principles, strengths, and limitations of these leading MSI techniques. Whether you’re focused on high-resolution imaging, ambient analysis, or detailed surface characterization, this article will provide the insights you need to make an informed choice.
Mass spectrometry imaging (MSI)
Mass spectrometry imaging (MSI), also known as imaging mass spectrometry, is an advanced molecular imaging technology capable of directly obtaining structural, quantitative, and spatial distribution information for numerous known and unknown endogenous metabolites or exogenous drugs from biological tissues. MSI offers distinct advantages, such as probe-free labeling, non-specific detection, and the simultaneous imaging of hundreds of metabolites in a single analysis. These features provide a novel approach to metabolomics research, addressing the limitations of techniques like gene amplification, staining, and labeling, which are unsuitable for metabolomics studies. Consequently, MSI has been widely applied in studying the spatiotemporal distribution of drugs, metabolites, peptides, and proteins in animal tissues, plant tissues, and even single cells.
The first MSI technology to emerge was Secondary Ion Mass Spectrometry (SIMS) in 1967, initially used for imaging and analyzing elements and small organic molecules. In 1987, a significant breakthrough occurred with the development of Matrix-Assisted Laser Desorption/Ionization (MALDI) technology, which enabled the measurement of proteins with molecular weights greater than 10 kDa. This advancement paved the way for the application of MALDI mass spectrometry in detecting biomolecules. In 1997, the integration of MALDI with Mass Spectrometry Imaging (MSI) for the first time allowed for the imaging of small molecules, peptides, and proteins in biological tissues, marking the beginning of a new era in MSI. Since then, the technology has continuously evolved, expanding its applications and becoming one of the most widely used and powerful analytical methods today.
The ion source is a critical component in mass spectrometry imaging. Based on variations in ion sources, MSI is typically categorized into the following types:
● Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging (MALDI-MSI)
● Secondary Ion Mass Spectrometry Imaging (SIMS)
● Desorption Electrospray Ionization Mass Spectrometry Imaging (DESI-MSI)
● Laser Ablation-Electrospray Ionization Mass Spectrometry Imaging (LA-ESI-MSI)
Each method offers unique advantages (see Table 1), with MALDI being the most widely applied. In addition to these core techniques, new ionization methods such as MALDI-2 (post-ionization), Matrix-Assisted Laser Desorption Electrospray Ionization (MALDESI), and Laser Ablation Inductively Coupled Plasma (LA-ICP) have recently emerged, contributing to the continued innovation of MSI applications. The introduction of Ion Mobility Spectrometry (IMS) has further enhanced the ability to differentiate isomeric compounds with varying collision cross-sections (CCS).
Table 1. Summary of Key Parameters for Commonly Used Mass Spectrometry Imaging Techniques (Yin et al., 2021)
_1737009749_WNo_1264d777.webp)
Matrix-Assisted Laser Desorption/Ionization (MALDI)
MALDI-MSI is the most commonly used MSI technique. The fundamental principle of MALDI-MSI involves depositing matrix molecules, which can absorb UV laser light at wavelengths of 337 nm or 355 nm, onto tissue sections via techniques such as air spraying or sublimation. These matrix molecules then interact with the analytes on the tissue surface, forming co-crystals. The excess matrix absorbs most of the laser energy, becoming excited or evaporating into the gas phase, thereby carrying the analytes into the mass spectrometer for detection. The process primarily relies on gas-phase proton transfer between the analyte and matrix molecules. Typically, the amount of matrix deposited far exceeds that of the analyte to ensure efficient desorption and ionization. Additionally, matrix molecules absorb the laser energy, preventing excess energy from directly affecting the analytes and causing fragmentation.
In the MALDI-MSI analysis process, the first step is selecting an appropriate sample preparation method based on the nature of the plant or animal tissue. For example, animal tissues (such as brain or kidney) and plant tissues (such as roots and stems) are typically frozen and sectioned to a thickness of approximately 5-20 μm, while leaf and petal samples are often prepared using imprint transfer. Next, the suitable MALDI matrix is selected based on the type and characteristics of the analytes. A laser beam is then used to desorb and ionize the sample at each sampling point. The tissue sample is moved using an XY two-dimensional platform, and the ions are separated and detected by the mass spectrometer, producing mass spectra that are linked to the spatial locations of the sample. Finally, the mass spectrometry data are matched to the corresponding two-dimensional spatial positions, and the MSI map is reconstructed.
_1737009813_WNo_1275d622.webp)
Figure 1. Schematic drawing of sample preparation process for MSI experiments (Ma and Fernández, 2022)
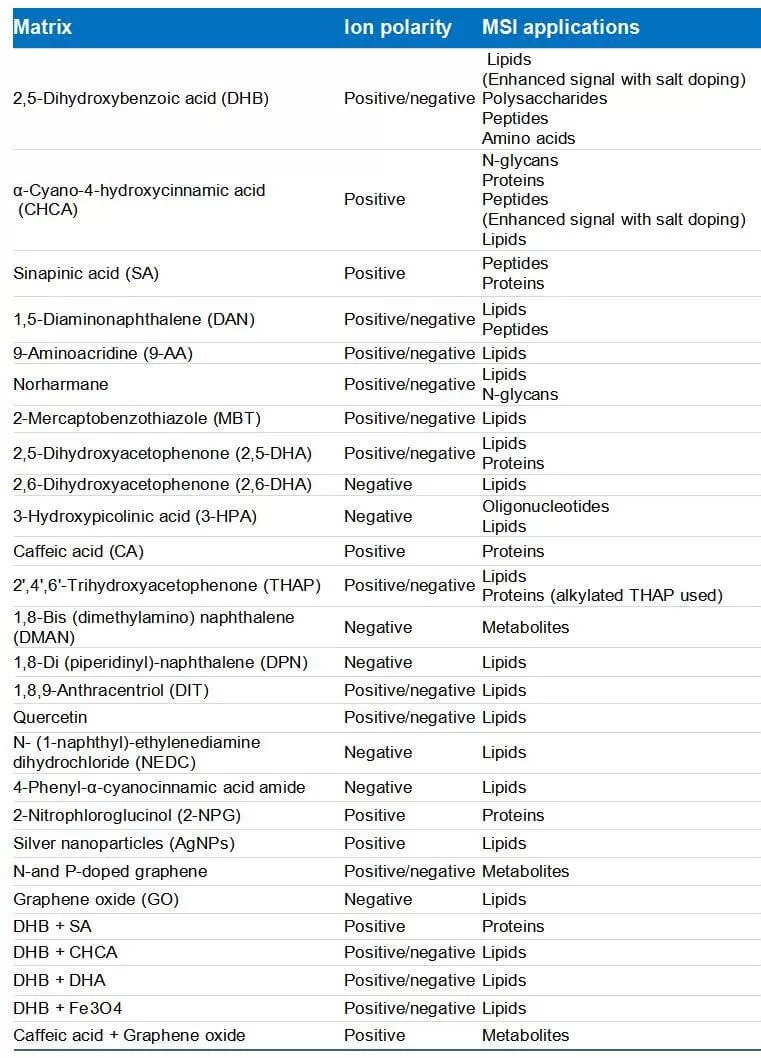
The matrix plays a vital role in the ionization process of MALDI. The choice of matrix depends on the experimental objectives, as different biomolecules are optimally ionized by different matrices. The physicochemical properties of the matrix compounds influence the quality of matrix deposition, and the choice of matrix can significantly impact ionization efficiency by producing varying crystal forms. In some cases, mixing two or more matrix compounds can enhance the ionization of specific molecules, such as lipids and oligosaccharides. Additionally, the development of novel matrices, such as nanoparticle matrices for MALDI-MSI, has become a key area of research. Table 2 shows some common matrices and binary matrix mixtures used in MALDI-MSI experiments.
The selection of a matrix deposition technique also plays a critical role in the quality of mass spectrometry images and the reproducibility of results. Currently, automated spraying and sublimation are the most commonly used deposition techniques. To achieve uniform matrix deposition, parameters such as solvent flow rate, drying gas flow rate, nozzle speed, temperature (spray nozzle temperature or sublimation chamber temperature), and humidity must be carefully optimized and controlled. In automated spraying, the water content in the matrix solution is an important factor, as it facilitates the diffusion of analytes on the surface. In contrast, sublimation is a dry matrix deposition technique that avoids such diffusion effects, although it may result in lower ionization efficiency for certain analytes. Furthermore, matrix-free MALDI-MSI techniques, also known as Surface-Assisted Laser Desorption/Ionization (SALDI), can eliminate interference from matrix ions, particularly for small molecules with low mass-to-charge ratios (m/z < 350). SALDI avoids molecular diffusion on the tissue surface during matrix deposition, promoting more precise compound localization, stronger ion signals, and broader compound coverage. In addition to SALDI, Nanoparticle-Assisted Laser Desorption/Ionization (NALDI) has also emerged as a matrix-free technique used in MSI studies of various tumor tissues.
Table 2. Matrix molecules and binary matrix systems used in different MALDI MSI experiments (Ma and Fernández, 2022)
Desorption Electrospray Ionization (DESI)
Desorption Electrospray Ionization (DESI) is a powerful ambient ionization technique used for mass spectrometry imaging (MSI). Unlike MALDI-MSI and SIMS, which operate under high-vacuum conditions, DESI-MSI enables ionization and imaging of analyte molecules at atmospheric pressure without requiring a matrix solution. This simplifies sample preparation, making it suitable for both small and large molecule analysis and enabling true in situ analysis. In 2004, the DESI source was introduced, operating on the principle that an extraction solvent, under the influence of nebulizer gas and voltage, forms an electrospray that sweeps across the sample surface at a specific angle. When the solvent contacts the surface, it quickly dissolves the analytes and forms charged droplets, which are directed into the mass spectrometer for detection. The ionization efficiency can be adjusted by changing the composition of the spray solvent. The proposed mechanism for DESI is referred to as "droplet picking," where the surface is wetted, and molecules are extracted.
Operating under ambient temperature and pressure, DESI-MSI eliminates the need for complex sample preparation or matrix assistance, thus preventing analyte loss due to poor matrix deposition and avoiding ion suppression from matrix ions. These advantages have led to its widespread use in imaging both plant and animal tissues. Various mass spectrometers, including Q-TOF, Orbitrap, and FTICR, have been successfully coupled with DESI for imaging applications. However, DESI-MSI also has certain limitations. For example, the composition of the electrospray solvent can result in differences in desorption and ionization efficiency for various analytes. Additionally, due to the diffusion of electrospray droplets upon reaching the tissue surface, DESI-MSI generally has lower spatial resolution, typically around 50-200 µm, making it more suitable for imaging large tissue samples.
Secondary Ion Mass Spectrometry (SIMS)
Secondary Ion Mass Spectrometry (SIMS) is a highly sensitive and high-resolution imaging technique that allows for the analysis of molecular and elemental composition on surfaces at the nanoscale. Compared to MALDI and DESI-MSI, SIMS is currently the method with the highest spatial resolution in mass spectrometry imaging. SIMS offers the highest spatial resolution (50-100 nm, as shown in Table 1), making it suitable for single-cell imaging. Unlike MALDI or DESI, which use ablation lasers or charged electrospray, SIMS utilizes a focused ion beam ("primary ion beam"), typically composed of metals, fullerenes, or gas clusters, to desorb and ionize molecules. Molecules from the tissue surface are ionized, generating secondary ions that are transferred to the mass analyzer for detection.
SIMS can be divided into dynamic and static types, based on the ion beam type and beam current. Dynamic SIMS typically uses high-energy sputtering ion beams, such as O- (positive ion mode) and Cs+ (negative ion mode) sources, to achieve ultra-high spatial resolution down to 50 nm. However, due to the large ion current, which is highly destructive, dynamic SIMS breaks all molecules into single-atom or multi-atom ions, making it suitable only for elemental imaging. Using elements to represent entire molecules for spatial localization often suffers from interference from background elements, preventing an accurate reflection of the original distribution of metabolites in cells. Furthermore, despite its unparalleled spatial resolution, dynamic SIMS remains limited in metabolomics and biomolecular imaging due to its inability to provide complete molecular information.
To address these limitations, static SIMS, which employs multi-atom ion beams and cluster ion beams, complements dynamic SIMS for molecular imaging. Static SIMS is suitable for single-cell imaging of molecules with molecular weights under 1000 Da, such as lipids, metabolites, small molecule drugs, and certain elements. While static SIMS can achieve sub-micron spatial resolution for elemental and molecular fragments, its relatively lower ionization efficiency means that its spatial resolution for intact molecules remains at the low-micron scale. As a result, its application in spatial metabolomics is still under development and requires further advancements.
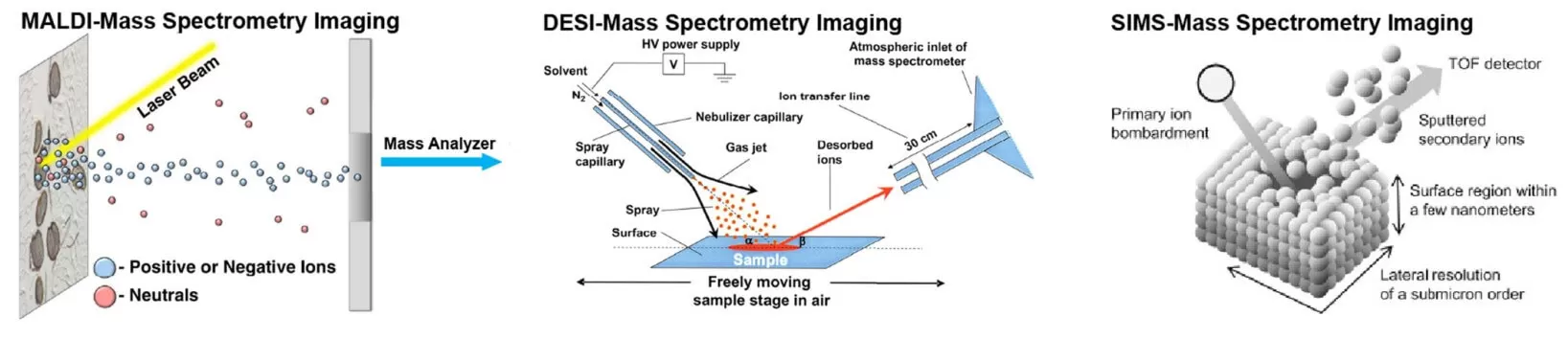
Figure 2. Schematic illustrations of the ionization steps in MALDI, DESI, and SIMS MSI experiments (Ma and Fernández, 2022)
Laser Ablation Electrospray Ionization (LA-ESI)
Laser Ablation Electrospray Ionization (LA-ESI) is a widely used mass spectrometry imaging (MSI) technique known for its high spatial resolution, high repetition rate, and high energy output. First introduced in 2007, the LA-ESI source operates by focusing and transmitting a 2.94 µm mid-infrared laser through a focusing lens or etched optical fiber. In LA-ESI, the laser irradiates the sample surface under ambient conditions, resonating with water molecules in the tissue and causing the compounds within the sample to be sputtered and evaporated. These vaporized molecules are then captured and ionized by the electrospray ionization (ESI) source, with the resulting ions directed into the mass spectrometer for analysis and imaging.
Compared to DESI-MSI, LA-ESI-MSI offers significantly higher spatial resolution, typically ranging from 30 to 300 µm. High-resolution imaging of Allium cepa epidermal cells has been achieved with a spatial resolution of 10-15 µm using an 800 nm femtosecond laser for non-resonant sputtering. Like DESI, LA-ESI’s advantages include direct analysis under atmospheric pressure without the need for a matrix and minimal sample preparation. However, the ionization process requires tissue samples to contain sufficient moisture to resonate with the far-infrared laser. While non-resonant femtosecond lasers can overcome this limitation, their high cost remains a consideration. Additionally, the technique's spatial resolution makes it best suited for broader analyses of plant cells and imaging of larger plant tissues. Moreover, LA-ESI is particularly effective for the ionization of polar lipids.
Other Mass Spectrometry Imaging Methods
In addition to the aforementioned methods, researchers have developed several newer ionization techniques for mass spectrometry imaging (MSI). Among these, AP-MALDI, Matrix-Assisted Laser Desorption Electrospray Ionization (MALDESI), and MALDI-2 have emerged as powerful imaging tools in recent years. These techniques have been widely applied for imaging cancer biomarkers, such as metabolites, lipids, glycans, and peptides, in tissues and single cells.
AP-MALDI-MSI operates on a principle similar to that of MALDI-MSI, where a matrix is sprayed onto the sample surface, allowing the matrix and sample compounds to co-crystallize before subsequent analysis. Unlike MALDI-MSI, which requires vacuum conditions for matrix deposition and ionization of compounds, AP-MALDI-MSI achieves ionization under atmospheric pressure using a "soft ionization" technique. Both matrix deposition and ionization occur under atmospheric pressure, which eliminates the vacuum waiting times associated with traditional MALDI-MSI.
Similarly to MALDI, MALDESI also requires a matrix to facilitate the desorption of molecules from the tissue surface. However, in MALDESI, the desorbed molecules are ionized by ESI solvents, and organic matrix compounds can be replaced by endogenous water or a thin ice layer deposited on the tissue surface. A key feature of MALDESI is its ability to generate multiply charged ions, making it particularly suited for imaging large biomolecules.
MALDI-2 involves a two-step post-ionization process, which includes ionization of matrix molecules induced by the primary MALDI laser pulse, followed by chemical ionization via a secondary laser pulse that transfers charge to the analyte. Under optimized experimental conditions, including both ionization modes and instrument parameters, MALDI-2 can enhance the ionization efficiency and sensitivity of various molecules by 2 to 3 orders of magnitude.
 LAESI, (B) MALDESI and (C) MALDI‐2 setups for_1737009924_WNo_918d723.webp)
Figure 3. Schematic illustrations of (A) LAESI, (B) MALDESI and (C) MALDI‐2 setups for MSI experiments (Ma and Fernández, 2022)
How to Choose the Right MSI Technique for Spatial Metabolomics
Selecting the most suitable mass spectrometry imaging (MSI) technique is essential for successful spatial metabolomics studies. MALDI, DESI, and SIMS each offer unique capabilities in terms of spatial resolution, sample preparation complexity, ionization efficiency, and analyte coverage. Your choice should be guided by the specific needs of your research—whether it’s high-resolution cellular imaging, ambient-condition analysis, or high-throughput metabolite profiling. Below is a comprehensive comparison to help you determine which MSI method aligns best with your experimental goals.
Comparison of MALDI, DESI, and SIMS in Spatial Metabolomics
| MSI Technique | Spatial Resolution | Ionization Environment | Sample Preparation | Analyte Coverage | Advantages | Limitations | Recommended Applications |
| MALDI (Matrix-Assisted Laser Desorption/Ionization) | 10–200 µm (typical) | Vacuum | Requires matrix, thin sectioning or imprint | Broad (proteins, lipids, metabolites) | High sensitivity, high-throughput, good for complex tissues | Requires matrix, may suppress low-mass analytes, vacuum conditions | Comprehensive metabolite imaging, tissue profiling in animals/plants |
| DESI (Desorption Electrospray Ionization) | 50–200 µm | Ambient | Minimal (no matrix needed) | Small molecules, lipids | Ambient analysis, minimal prep, in situ imaging | Lower resolution, ion suppression possible, sensitive to solvent composition | In situ tissue imaging, plant research, clinical applications requiring fast turnaround |
| SIMS (Secondary Ion Mass Spectrometry) | 50 nm – few µm | Vacuum | High vacuum, no matrix, rigid prep | Mainly small molecules, elements | Ultra-high spatial resolution (nanoscale), label-free | Destructive, low molecular coverage, limited to low MW compounds | Single-cell or subcellular imaging, elemental mapping, nanoscale surface analysis |
Reference
Ma X, Fernández FM. Advances in mass spectrometry imaging for spatial cancer metabolomics. Mass Spectrometry Reviews, (2022); e21804. https://doi.org/10.1002/mas.21804
Yin ZB, Huang WJ, Wu XZ and Yan SJ. Spatially Resolved Metabolomics: Progress and Challenges. Biotechnology Bulletin, (2021); 37(1):32-51. https://doi.org/10.13560/j.cnki.biotech.bull.1985.2020-1374
Read more
- LC-MS VS GC-MS: What's the Difference
- LC vs. HPLC vs. UHPLC: Tracing the Evolution of Chromatographic Techniques
- Mastering Chromatography: Everything You Need to Know
- DIA Proteomics vs DDA Proteomics: A Comprehensive Comparison
- Multi-Omics Association Analysis Series
- Omics Data Processing Series
- Omics Data Analysis Series
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


