Metabolomics in Chronic Kidney Disease
 We are very pleased to share a groundbreaking Metabolomics study on chronic kidney disease (CKD) titled “Metabolome profiling by widely-targeted metabolomics and biomarker panel selection using machine-learning for patients in different stages of chronic kidney disease” in the Chinese Chemical Letters Journal. This research highlights the use of MetwareBio's TM Widely-Targeted Metabolomics method, leveraging LC-MS technology to analyze plasma samples from healthy controls and CKD patients at various stages. The study identified 1,431 metabolites, with significant findings that include a machine learning-derived predictive model for CKD staging, showcasing an impressive AUC of 0.99.
We are very pleased to share a groundbreaking Metabolomics study on chronic kidney disease (CKD) titled “Metabolome profiling by widely-targeted metabolomics and biomarker panel selection using machine-learning for patients in different stages of chronic kidney disease” in the Chinese Chemical Letters Journal. This research highlights the use of MetwareBio's TM Widely-Targeted Metabolomics method, leveraging LC-MS technology to analyze plasma samples from healthy controls and CKD patients at various stages. The study identified 1,431 metabolites, with significant findings that include a machine learning-derived predictive model for CKD staging, showcasing an impressive AUC of 0.99.
Metabolome Profiling Using Widely-Targeted Metabolomics
WT-Met(Widely-Targeted Metabolomics) is an ultra-sensitive, wide-coverage metabolomics method that can simultaneously identify and quantify complex metabolites in human blood samples. This method combines the advantages of both targeted and untargeted metabolomics, enabling large-scale identification and accurate quantification of thousands of metabolites. The process begins with high-resolution mass spectrometry to obtain fragment information of compounds in mixed samples, which are then matched against a local database containing 2,000 standards to create a project-specific database after deduplication. Next, the method uses triple quadrupole mass spectrometry in MRM mode to accurately quantify compounds in the samples. Finally, TOF-MS and MRM-IDA-EPI are employed to assist in the qualitative analysis, enhancing the accuracy of unknown metabolite identification. The detailed workflow is as follows.

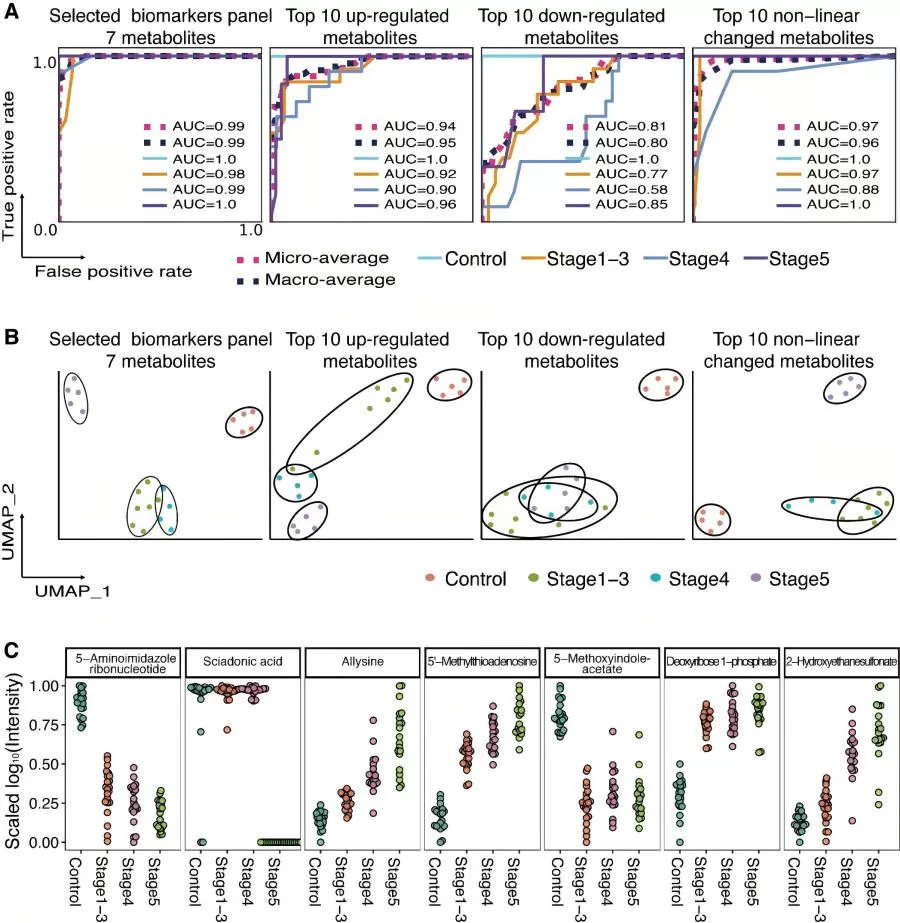
Machine Learning in Biomarker Selection and Predictive Modeling
Metabolomic data is characterized by high dimensionality, high correlation, complexity, disorder, and abundance variations spanning several orders of magnitude. Traditional statistical methods are inefficient in handling such data, making it difficult to uncover key patterns and trends, and often resulting in limited insights. In contrast, machine learning possesses powerful capabilities for automatically learning from complex data, effectively identifying differential metabolites, and constructing predictive models to enhance prediction accuracy.
In this study, the authors first employed feature selection strategies in machine learning to identify a set of 7 metabolites as biomarkers. These biomarkers were then used to fit various machine learning algorithms, with grid search and cross-validation employed to determine the optimal parameter combinations for each algorithm. Finally, the best-performing algorithm was selected as the predictive model based on evaluation metrics such as accuracy and F1 score. This model achieved a macro-AUC and micro-AUC of 0.99 on the test set.
At MetwareBio lab in Boston, we provide comprehensive metabolomics and proteomics analysis services to enhance your research endeavors. Contact us today to learn how our state-of-the-art techniques can support your studies and drive scientific discoveries.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


