Metabolomics Biomarker Screening Process
Metabolomics Biomarker Series Article Catalog:
- Unlocking Biomarkers: A Guide to Vital Health Indicators
- Metabolomics and Biomarkers: Unveiling the Secrets of Biological Signatures
- Choosing the Right Study Design for Metabolomics Biomarker Discover
The more accessible clinical study type focusing on the metabolome is the observational cross-sectional study, i.e., to identify metabolic markers associated with a certain disease through the metabolome, followed by the diagnosis of the disease or basic mechanistic research. Therefore, in this section, we will present the design scheme of the metabolic marker screening process for cross-sectional studies.
To begin with, let's look at two cases:

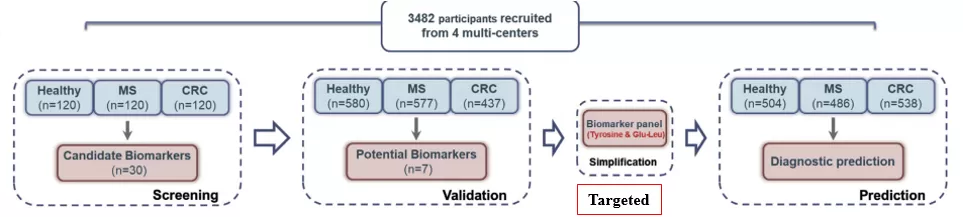
① Discovery phase 1- High-throughput metabolomics assays were performed on 360 subjects and 30 differential metabolites were screened;
② Discovery phase 2- High-throughput detection was performed on 1594 subjects with seven differential metabolites shared with discovery phase 1 being screened; two potential biomarkers were subsequently derived by binary logistic regression analysis;
③ Modeling phase - A diagnostic model was constructed based on targeted detection against two biomarkers in 900 subjects (from discovery phase 2 );
④ Validation phase - The diagnostic efficacy of the model was validated by targeted detection against two biomarkers in 1528 subjects.

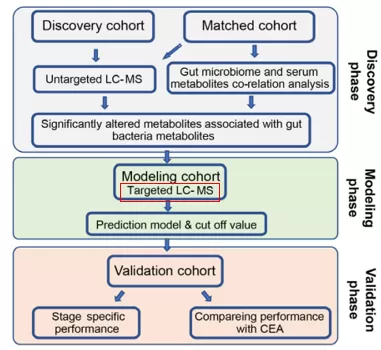
- In the discovery phase, high-throughput metabolomics detection was performed on serum samples from 92 subjects, with 44 serum-matched fecal samples being selected for macro-genomic detection. A total of 322 metabolites related to the gut microbiome were identified in the conjoint analysis, and eight metabolites were screened and identified using the LASSO algorithm;
- In the modeling phase, the abundance of eight metabolites was measured in 72 normal individuals and 120 patients with colorectal anomalies using a targeted approach, and a predictive model was generated using logistic regression;
- In the validation phase, the predictive performance of this model was evaluated by performing targeted detection of 8 metabolites in the validation cohort consisting of 103 patients with colorectal anomalies and 53 healthy individuals.
It is not difficult to notice from the above two typical cases that the screening process of metabolomics biomarkers usually consists of three phases: ① discovery phase, in which candidate biomarkers are screened by high-throughput metabolome screening in the discovery cohort; ② modeling phase, in which screened biomarkers are detected in the modeling cohort by a targeted approach and a discriminative model is constructed; ③ validation phase, in which biomarkers are target-detected in the validation cohort to validate the model's discriminatory efficacy.

So, which phase does the discovery set, training set, validation set, and test set we often read about in metabolome articles refer to? These terms are actually derived from the process of machine learning. In machine learning, samples are generally divided into three separate parts: the training set, the validation set, and the test set.
- Training set - the data samples used for model fitting.
- Validation set - a separate set of samples set aside during model training that can be used to tune the hyperparameters of the model and for initial evaluation of the model's capabilities.
- Test set - is used to evaluate the generalization ability of the final model. However, it cannot be used as a basis for algorithm-related choices such as tuning and selecting features.
If we map these concepts to the metabolic marker screening process, the population in discovery phase 1 corresponds to the training set, which is also referred to as the discovery set in some articles; the population in discovery phase 2 corresponds to the validation set; the population in the validation phase corresponds to the test set; and the population in the modeling phase can either originate from the validation set or from the test set ( independent of the population in the validation phase).
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


