Isotope Labeling Techniques: A Comprehensive Guide and Emerging Applications
Introduction to Isotope Labeling
Isotope labeling, a cornerstone of modern scientific research, leverages the unique physical properties of isotopes (e.g., radioactive decay or mass differences) to trace molecular dynamics in biological, chemical, and physical processes. By replacing specific atoms in a compound with their isotopic counterparts, scientists gain unparalleled insights into pathways, metabolic fluxes, and reaction mechanisms. This blog delves into the principles, methodologies, and cutting-edge applications of isotope labeling, highlighting its transformative role across disciplines.
Types of Isotopes and Their Core Principles
1) Types of Isotopes
- Radioactive Isotopes (³H, ¹⁴C, ³²P, ³⁵S): Emit detectable radiation (e.g., β-particles) and are tracked via scintillation counters or autoradiography.
- Stable Isotopes (¹³C, ¹⁵N, ²H, ¹⁸O): Non-radioactive, analyzed by mass spectrometry (MS) or nuclear magnetic resonance (NMR) based on mass shifts or isotopic displacement.
2) Labeling Strategies
- Single Labeling: Targets one compound (e.g., [¹³C]-glucose).
- Parallel Labeling: Uses multiple isotopes (e.g., [¹³C]-glucose + [²H]-water) to reduce biological variability and enhance data robustness.
3) Fundamental Principles
- Chemical Equivalence: Isotopes share identical electronic configurations and reactivity (e.g., ¹²C vs. ¹⁴C-labeled leucine in protein synthesis).
- Physical Detectability:
- Radioactive isotopes: Quantified via radiation intensity (e.g., ³²P in DNA hybridization).
- Stable isotopes: Detected via MS (mass differences) or NMR (isotopic shifts).
- Isotope Dilution: Enables absolute quantification using labeled internal standards (e.g., SILAC for proteomics).
Methodologies and Experimental Workflows for Isotope Labeling
1) Radioactive Isotope Labeling
Workflow:
- Label Selection: Match isotopes to target molecules (e.g., ³⁵S for methionine in proteins).
- Sample Incorporation: Introduce isotopes into biological systems (cell cultures, animal models).
- Detection: Autoradiography (Southern blot) or liquid scintillation counting.
2) Stable Isotope Labeling
Metabolic Flux Analysis (¹³C-MFA):
- Labeled Substrates: Use ¹³C-glucose to trace carbon flow.
- Isotopomer Analysis: MS profiles metabolic intermediates to model flux networks.
SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture):
- "Heavy" Media: Incorporate ¹³C/¹⁵N-arginine/lysine into cell cultures.
- Quantitative Proteomics: MS compares labeled vs. unlabeled protein ratios for precise quantification.
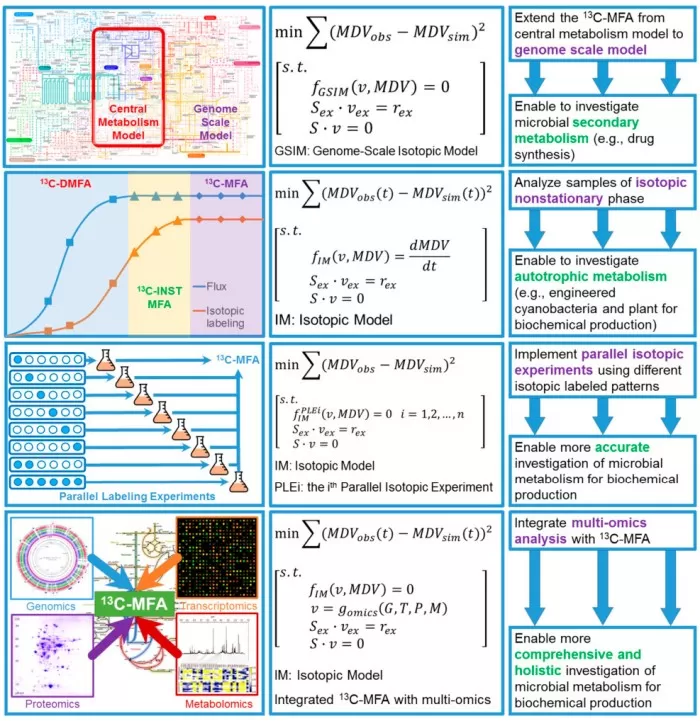
¹³C-MFA workflow or SILAC-based proteomics pipeline
3) Hybrid Techniques
- Fluorescence-Isotope Dual Labeling: Combines FITC (fluorescence) with ³H labeling for spatial and metabolic tracking.
- Isotope-Coded Affinity Tags (ICAT): Uses ²H/¹³C tags to enrich low-abundance proteins for enhanced MS sensitivity.
Key Isotopes and Their Applications
|
Isotope |
Type |
Applications |
|
³H |
Radioactive |
DNA/RNA synthesis tracking (³H-thymidine) |
|
¹⁴C |
Radioactive |
Organic metabolism studies (¹⁴C-glucose) |
|
³²P |
Radioactive |
Nucleic acid labeling (³²P-dCTP in hybridization assays) |
|
³⁵S |
Radioactive |
Protein synthesis monitoring (³⁵S-methionine) |
|
¹³C |
Stable |
Metabolic flux analysis (¹³C-MFA) |
|
¹⁵N |
Stable |
Protein structural studies (¹⁵N-amino acids in NMR) |
|
¹⁸O |
Stable |
Oxygen source tracing in photosynthesis (H₂¹⁸O) |
|
²H |
Stable |
Lipid membrane dynamics (²H-palmitic acid in deuterium NMR) |
Emerging Applications of Isotope Labeling
1) Synthetic Biology
Case Study: Optimizing microbial cell factories using ¹³C-labeled substrates to enhance biofuel production. Researchers at MIT engineered E. coli strains with ¹³C-glucose, mapping carbon flux bottlenecks and increasing ethanol yield by 40%.
2) Clinical Medicine
Drug Pharmacokinetics: ¹⁴C-labeled anticancer drugs (e.g., paclitaxel) track absorption and clearance in vivo, guiding personalized dosing regimens.
Biomarker Discovery: SILAC-based proteomics identified serum amyloid A (SAA) as a biomarker for early-stage lung cancer in a 2023 Nature study.
3) Environmental Science
Pollution Tracking: ¹⁵N-labeled nitrates traced agricultural runoff in the Mississippi River Basin, revealing fertilizer overuse hotspots.
Carbon Sequestration: ¹³CO₂ labeling quantified carbon fixation rates in reforested Amazonian soils, aiding climate mitigation strategies.
4) Geochronology
¹⁴C Dating: Accurately dated Neanderthal fossils (±40 years), refining human evolutionary timelines.
Isotope labeling bridges biology, chemistry, and physics, driving innovations from Calvin cycle elucidation to multi-omics integration. Advances in miniaturized detectors, AI-driven data analysis, and multi-isotope labeling will unlock new frontiers in precision medicine, sustainable biomanufacturing, and environmental monitoring. Whether unraveling metabolic networks or combating climate change, isotope labeling remains an indispensable tool for decoding nature’s complexity.
References
Du Q, Yu X, Jia K, Qu Y, Han J, Sun J, Shen D, Liu H, Nie Z. Chemoselective and laser cleavable probes for in situ protein lipoylation detection by laser desorption/ionization mass spectrometry. Chem Sci. 2025 Feb 10;16(11):4860-4865. doi: 10.1039/d4sc05553e.
Li X, Hu H, Yang M, Laskin J. A Low-Cost, High-Resolution Thermoplastic Microfluidic Probe for Mass Spectrometry Imaging of Biological Tissue Samples. Anal Chem. 2025 Feb 18;97(6):3207-3212. doi: 10.1021/acs.analchem.4c06087.
Bocoș-Bințințan V, Bocoș-Bințințan PF, Rozsypal T, Beldean-Galea MS. Trace Detection of Di-Isopropyl Methyl Phosphonate DIMP, a By-Product, Precursor, and Simulant of Sarin, Using Either Ion Mobility Spectrometry or GC-MS. Toxics. 2025 Jan 28;13(2):102. doi: 10.3390/toxics13020102.


