How Does SDS-PAGE Work? A Comprehensive Guide
1. Introduction to Electrophoresis
Electrophoresis is a fundamental technique used to separate proteins, nucleic acids, organic compounds, and even inorganic ions based on their mobility in a gel matrix under an electric field. Among various gel types, polyacrylamide gel is widely favored for protein separation due to its tunable pore size (matching protein dimensions), chemical stability, pH/temperature resistance, and high reproducibility. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) stands out for its simplicity and reliability in determining protein molecular weights, detecting specific proteins, and identifying bacterial strains.
2. Principles of SDS-PAGE
2.1 Composition of Polyacrylamide Gel
Polyacrylamide gel is formed through the polymerization of acrylamide and the cross-linker N,N'-methylenebisacrylamide, catalyzed by ammonium persulfate (APS) and N,N,N',N'-tetramethylethylenediamine (TEMED). This creates a three-dimensional network with pores that separate proteins based on their molecular weights (MW) and charge.
2.2 Role of SDS
SDS, an anionic detergent, binds to proteins at a ratio of ~1.4 g SDS per gram of protein in the presence of reducing agents (e.g., β-mercaptoethanol or DTT). This disrupts hydrogen and hydrophobic bonds, denaturing proteins into linear SDS-protein complexes. These complexes carry a uniform negative charge, masking inherent charge differences. Consequently, protein migration in SDS-PAGE depends solely on MW—smaller proteins migrate faster through the gel matrix.
2.3 Gel Structure and Electrophoresis Process
An SDS-PAGE gel consists of two layers:
- Stacking Gel (pH 6.8): Low acrylamide concentration (~5%) compresses protein samples into sharp bands during initial electrophoresis.
- Separating Gel (pH 8.8): Higher acrylamide concentration (~10-15%) separates proteins by size.
Key Steps in Electrophoresis:
1. Sample Loading: Proteins mixed with SDS and reducing agents are heat-denatured (95°C, 5 min) to ensure complete linearization.
2. Electrophoresis Setup: A Tris-glycine-SDS buffer (pH 8.3) facilitates migration.
3. Run Conditions:
- Stacking Phase: 80 V to concentrate proteins into thin bands.
- Separating Phase: 120 V to resolve proteins by size.
- Cooling (e.g., ice bath) is recommended to prevent overheating.
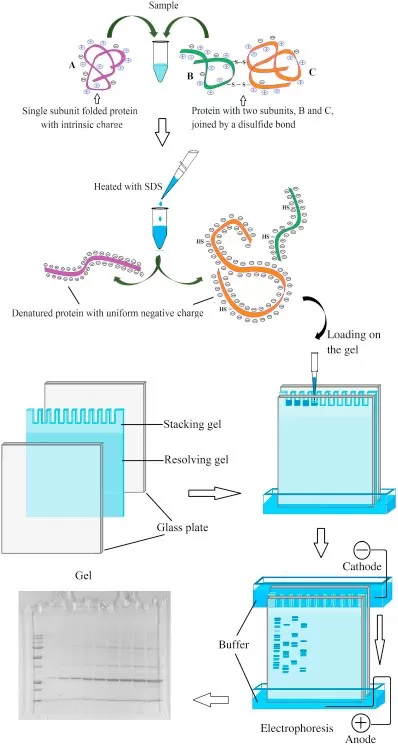
Schematic of SDS-PAGE gel structure and protein migration
3. Step-by-Step Experimental Protocol
3.1 Reagent Preparation
10% Separating Gel (pH 8.8):
|
Component |
Volume (10 mL) |
|
30% Acrylamide/Bis Mix |
3.3 mL |
|
1.5 M Tris-HCl (pH 8.8) |
2.5 mL |
|
10% SDS |
100 μL |
|
Deionized Water |
3.9 mL |
|
10% APS |
50 μL |
|
TEMED |
5 μL |
5% Stacking Gel (pH 6.8):
|
Component |
Volume (5 mL) |
|
30% Acrylamide/Bis Mix |
0.83 mL |
|
1.0 M Tris-HCl (pH 6.8) |
0.63 mL |
|
10% SDS |
50 μL |
|
Deionized Water |
3.4 mL |
|
10% APS |
25 μL |
|
TEMED |
5 μL |
Safety Note: Acrylamide is neurotoxic—wear gloves and work in a fume hood.
3.2 Gel Casting (Mini-PROTEAN System)
1) Separating Gel:
- Assemble glass plates, mix components (excluding APS/TEMED), degas, then add APS/TEMED.
- Pour gel solution, overlay with isopropanol, and polymerize for 20-30 min.
2) Stacking Gel:
- Remove isopropanol, prepare stacking gel mixture, degas, add APS/TEMED, and insert a comb. Polymerize for 15-20 min.
3) Sample Preparation
- 2× Laemmli Buffer: 4% SDS, 20% glycerol, 0.004% bromophenol blue, 100 mM Tris-HCl (pH 6.8), 10% β-mercaptoethanol (added fresh).
- Procedure: Mix protein sample with buffer, boil (95°C, 5 min), cool on ice, and centrifuge.
- Loading: 20-50 μg/well for Coomassie staining; 1-10 μg/well for silver staining.
4) Electrophoresis
Running Buffer: 25 mM Tris, 192 mM glycine, 0.1% SDS (pH 8.3).
Conditions:
- Stacking phase: 80 V until dye front enters separating gel.
- Separating phase: 120 V until dye reaches the gel bottom (~60-90 min).
5) Staining and Destaining
Coomassie Brilliant Blue Protocol:
(1) Fixation: 40% ethanol + 10% acetic acid, 30 min.
(2) Staining: 0.1% Coomassie R-250 (in 40% ethanol + 10% acetic acid), 1-2 hr.
(3) Destaining: 10% ethanol + 7% acetic acid until background clears.
Silver Staining (High Sensitivity):
(1) Fixation: 50% ethanol + 5% acetic acid, 30 min.
(2) Sensitization: 0.02% sodium thiosulfate, 1 min.
(3) Staining: 0.1% silver nitrate (+ formaldehyde), 20 min.
(4) Development: 2% sodium carbonate (+ formaldehyde) until bands appear.
(5) Termination: 5% acetic acid, 10 min.
4. Determining Protein Molecular Weight
SDS-PAGE enables MW estimation by comparing sample migration to a pre-stained protein ladder (e.g., 10–250 kDa). A semi-log plot of MW vs. relative migration distance (Rf) is used to interpolate unknown protein sizes.
5. Interpreting SDS-PAGE Results
Post-electrophoresis, proteins are visualized via staining. Coomassie is cost-effective for abundant proteins, while silver staining offers 100x higher sensitivity. Advanced methods (e.g., fluorescent dyes) enhance compatibility with automated proteomic platforms.
6. Gel Storage Guidelines
- Fresh gels can be stored in water at 4°C for up to 1 week.
- Stained gels should be imaged promptly to prevent band diffusion.
7. Troubleshooting Common Issues
|
Issue |
Possible Cause |
Solution |
|
Smearing/Streaking |
Incomplete denaturation |
Extend boiling time; add protease inhibitors |
|
Vertical Streaks |
Air bubbles in gel |
Degas gel solution; tap plates gently |
|
Aberrant Migration |
Uneven SDS binding |
Use fresh DTT and sample buffer |
|
Failed Polymerization |
Degraded APS/TEMED |
Prepare fresh APS (store ≤1 week at 4°C) |
Conclusion
SDS-PAGE remains a cornerstone technique in molecular biology for protein analysis. Its ability to separate proteins by size with high resolution, coupled with straightforward protocols, makes it indispensable in research and diagnostics. By mastering the principles and troubleshooting strategies outlined here, researchers can achieve reliable and reproducible results.


