Maximizing Proteomic Potential: Top Software Solutions for MS-Based Proteomics
1. Introduction to MS-Based Proteomics Software
Bottom-up proteomics techniques can be categorized into three primary types: Data Dependent Acquisition (DDA), Data Independent Acquisition (DIA), and Parallel Reaction Monitoring (PRM). Key methodologies include MS1-based label-free quantification, TMT/iTRAQ/SILAC labeled quantification (DDA), DIA quantification, and targeted quantification using PRM. The commonly used proteomics analysis process includes protein extraction, reduction, alkylation, digestion, and purification, after which the resulting peptides are loaded into an LC-MS/MS system. Following data acquisition, the mass spectrometer produces a dataset that includes mass-to-charge ratios, intensity, retention time, and, when applicable, ion mobility (as seen in Bruker’s TIMS-TOF). This generates mass spectra for tens of thousands to over a hundred thousand peptides or fragments, resulting in highly complex data.
To navigate this complexity, a variety of software tools are used to analyze the mass spectra and facilitate the identification and quantification of peptides and proteins. In this blog, we will explore some of the most widely used software solutions in mass spectrometry-based proteomics, including SEQUEST, Mascot, Proteome Discoverer, MaxQuant, FragPipe, Skyline, DIA-NN, and Spectronaut, highlighting their functionalities and applications in protein identification and quantification.
2. SEQUEST: A Pioneer in Protein Identification
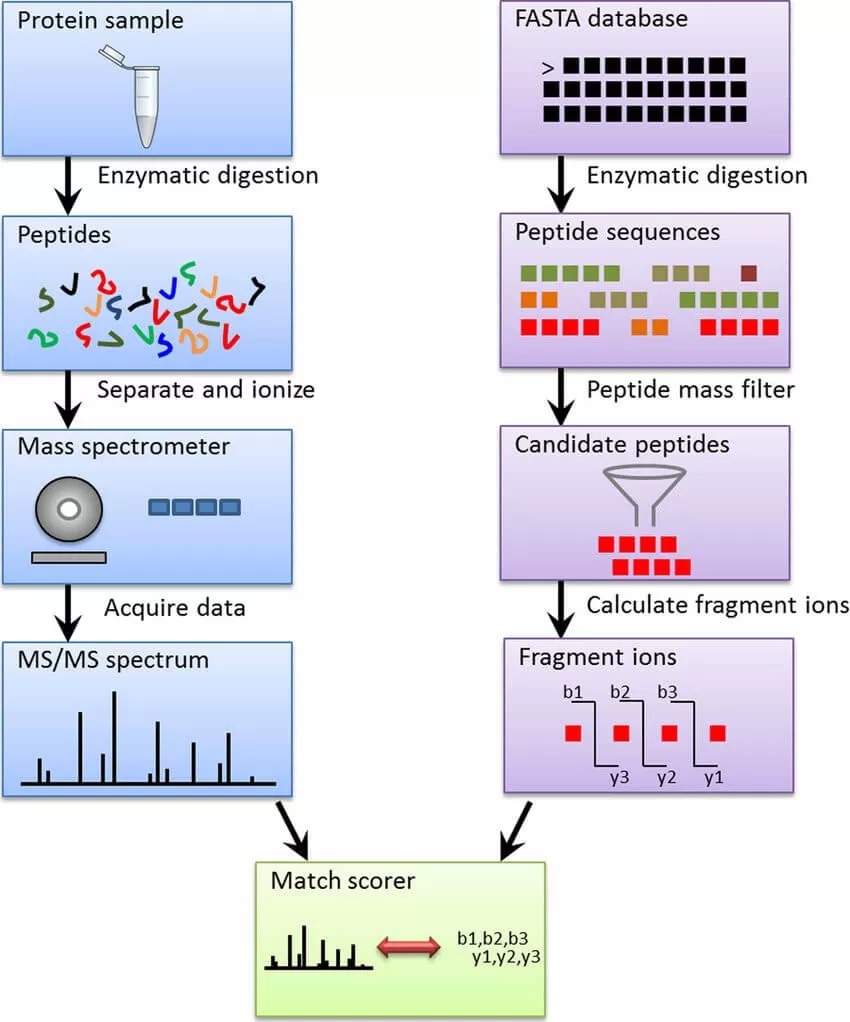
1) Introduction: SEQUEST is one of the pioneering database search engines for parsing LC-MS/MS data, developed by John Yates, a prominent figure in proteomics, in 1994. This software identifies each tandem mass spectrum individually by evaluating protein sequences from a database to generate a list of potential peptides. The intact mass of the peptide, as determined from the mass spectrum, guides SEQUEST in identifying candidate peptide sequences that are comparable to the observed peptide ion mass. For each candidate, SEQUEST generates a theoretical tandem mass spectrum and uses cross-correlation to compare these theoretical spectra with the observed mass spectrum. The candidate sequence that best matches the theoretical tandem mass spectrum is reported as the most likely identification for that spectrum (Fig. 1).
2) Range of Application: SEQUEST is designed specifically for the identification of peptides and proteins using DDA mode LC-MS/MS data.
3) Acquisition: SEQUEST is commercial software and is now integrated into the Proteome Discoverer™ platform.
3. Mascot: Widely Used in Peptide Sequence Databases
1) Introduction: Mascot is a widely used software search engine that identifies proteins from peptide sequence databases using mass spectrometry data. Developed by Matrix Science, Mascot employs a probabilistic scoring algorithm adapted from the MOWSE algorithm for protein identification. It is freely accessible for smaller data sets on the Matrix Science website. A license is required for in-house use, allowing for the integration of additional features.
2) Range of Application: Mascot is versatile and can be utilized for peptide and protein identification, iTRAQ/TMT quantification, top-down protein identification, and peptide mass fingerprinting using DDA tandem mass data from a variety of mass spectrometer manufacturers. The Mascot Distiller provides a single graphical user interface for native (binary) data files from platforms such as Agilent, AB Sciex, Bruker, Shimadzu, Thermo, and Waters. It processes raw data into high-quality, de-isotoped peak lists, identifies proteins through database searches and de novo sequencing, and quantifies proteins using isobaric, isotopic, and label-free methods. The Mascot Daemon allows for the automation of all steps. Distiller is available for free in read-only mode, serving as a project viewer for sharing search and quantitation results with colleagues.
3) Acquisition: Users can access the Mascot server at its official website. While Mascot is commercial software, a free online version is available for small datasets.
4. Proteome Discoverer: All-in-One Data Analysis Platform
1) Introduction: Proteome Discoverer (PD) is a comprehensive data analysis platform designed for qualitative and quantitative proteomics research, developed by Thermo Fisher. It is primarily compatible with the Orbitrap series of mass spectrometers, making it one of the most popular software solutions for identifying and quantifying DDA data in proteomics. As an all-in-one data analysis platform, PD integrates various relevant tools, including the Mascot, Comet, and SEQUEST search engines, as well as the Percolator algorithm for assessing and filtering search results. The latest version of PD features an intelligent search algorithm called CHIMERYS™, which enhances identification depth by approximately 12%. While the new version can also process DIA data from Astral or other Orbitrap-type mass spectrometers, its performance is not as robust as that of dedicated DIA search engines like DIA-NN and Spectronaut.
2) Range of Application: PD supports a range of applications, including LC-MS/MS data identification, label-free quantification in DDA mode, iTRAQ/TMT/SILAC labeled quantification, and label-free quantification in DIA mode for versions 3.0 and above.
3) Acquisition: PD is commercial software, and a license must be purchased for long-term use. Users can access a trial version of PD through the download portal at this webpage.
5. MaxQuant: Comprehensive Computational Proteomics
1) Introduction: MaxQuant (MQ) is a globally recognized platform for computational proteomics, developed by Dr. Jürgen Cox at the Max Planck Institute of Biochemistry in 2008. It allows researchers worldwide to perform highly precise analyses of their mass spectrometry data, online and free of charge. The MaxQuant suite comprises a set of algorithms for peak detection, peptide scoring, mass calibration, database searches for protein identification, and protein quantification, providing summary statistics. An FDR-controlled algorithm called "matching between runs" facilitates MS/MS-free identification of MS features across the entire dataset, enhancing the number of quantified proteins per sample. To achieve accurate and robust proteome-wide quantification through label-free approaches, MQ has introduced a normalization procedure called MaxLFQ, which is compatible with any peptide or protein separation prior to LC-MS analysis and has become widely adopted.
2) Range of Application: MaxQuant is designed for analyzing large-scale mass spectrometric data sets and supports all major labeling techniques, including SILAC, Di-methyl, TMT, and iTRAQ, as well as label-free and DIA (maxDIA) quantification. It can process measured spectra from various vendors, including Thermo Fisher Scientific, Bruker Daltonics, AB Sciex, and Agilent Technologies. Its user-friendly interface enables researchers to analyze complex data sets on standard desktop machines.
3) Acquisition: MaxQuant is written in C# and is freely available for download at https://www.biochem.mpg.de/6304115/maxquant. The download includes the Andromeda search engine and the Viewer module. The search engine comes preconfigured with a wide range of modifications, labels, proteases, and databases, while additional configurations can be customized for specialized studies. The Viewer module allows for inspection of both unprocessed and processed raw data. For statistical analysis of MaxQuant output, the Perseus framework is recommended.
6. FragPipe: Cutting-Edge Open Search Engine
1) Introduction: FragPipe is a newly developed comprehensive computational platform powered by MSFragger, an ultrafast proteomic search engine adept at both conventional and “open” peptide identification (with wide precursor mass tolerance). FragPipe integrates Percolator and the Philosopher toolkit for downstream post-processing of MSFragger search results, including PeptideProphet, iProphet, and ProteinProphet for FDR filtering, label-based quantification, and multi-experiment summary report generation. The platform also features the MSBooster module for deep-learning-based rescoring of peptide identifications, along with Crystal-C and PTM-Shepherd to assist in interpreting open search results. FragPipe includes additional modules like TMT-Integrator for TMT/iTRAQ quantification, IonQuant for label-free quantification with FDR-controlled match-between-run (MBR) functionality, EasyPQP for spectral library building, and MSFragger-DIA, DIA-Umpire SE, and diaTracer modules for direct library-free analysis of DIA data.
2) Range of Application: FragPipe is built with a Java Graphical User Interface (GUI) but can also be run in command-line mode on Windows, Linux, or in cloud environments. It processes a wide range of quantitative proteomics data, including label-free, TMT, iTRAQ, SILAC, and DIA. FragPipe supports various file formats, including .mzml, Thermo’s .raw, Bruker’s .d, and .mgf.
3) Acquisition: FragPipe is an open-source platform available for free at https://fragpipe.nesvilab.org/. For more information, please visit the homepage and refer to the associated publications.
7. Skyline: Free Targeted Proteomics Software
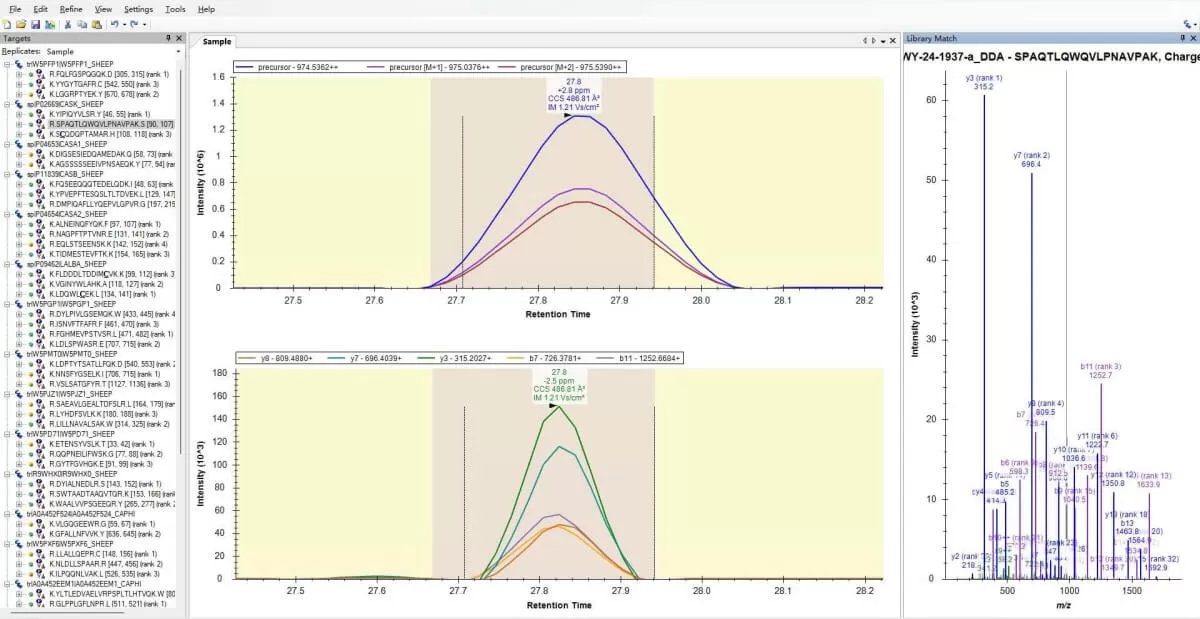
1) Introduction: Skyline is a freely available, open-source Windows application designed for building and analyzing Selected Reaction Monitoring (SRM), Multiple Reaction Monitoring (MRM), Parallel Reaction Monitoring (PRM), Data Independent Acquisition (DIA/SWATH), and DDA quantitative methods. Its standout feature is its visual design, which allows users to view each MS/MS peptide spectrum match alongside the Extracted Ion Current (XIC) plot. This visualization aids in evaluating and excluding low-quality identifications and quantifications. Additionally, Skyline displays various peptide and fragment characteristics, such as abundance and retention time, providing comprehensive insights into peptide behavior across all samples. The Skyline homepage offers numerous step-by-step tutorials, making it easy for users to navigate the software effectively.
2) Range of Application: Skyline is particularly well-suited for targeted proteomic data analysis, although it also accommodates label-free and DIA proteomics quantification.
3) Acquisition: Skyline software and tutorials are freely available on the official website.
8. DIA-NN: Leading Tool for DIA Proteomics
1) Introduction: DIA-NN is a universal software suite tailored for processing data-independent acquisition (DIA) proteomics data. First published in Nature in 2019, DIA-NN utilizes deep neural networks and innovative quantification and signal correction strategies to enhance the performance of DIA proteomic applications. It is particularly advantageous for high-throughput studies due to its speed and ability to deliver deep and confident proteome coverage, especially when paired with fast chromatographic methods. For diaPASEF data generated by timsTOF mass spectrometers, DIA-NN has developed algorithms for two-dimensional peak picking. DIA-NN supports both library-based and library-free DIA data generated by Thermo Fisher, Sciex, and Bruker. When operating in library-free mode, DIA-NN generates a predicted spectra library, eliminating the need for a project-specific DDA spectra library.
2) Range of Application: DIA-NN is designed for processing data-independent acquisition (DIA) proteomics data. It supports various formats, including Sciex .wiff, Bruker .d, Thermo .raw, .mzML, and .dia. DIA-NN is compatible with both Windows and Linux operating systems and is available for free.
3) Acquisition: DIA-NN can be downloaded from GitHub at https://github.com/vdemichev/DiaNN. The GitHub homepage also provides detailed instructions for installation and usage.
9. Spectronaut: DIA Data Analysis at Scale
1) Introduction: Spectronaut® is a commercial software package designed for analyzing data-independent acquisition (DIA) proteomics experiments. It can quantitatively profile hundreds to thousands of proteins in a single experiment, making it suitable for large studies that involve multiple conditions and replicates, with the capacity to analyze tens of thousands of LC-MS runs.
2) Range of Application: Spectronaut® is specifically tailored for processing data-independent acquisition proteomics data. It supports both library-based and direct DIA datasets, accommodating various formats such as Sciex .wiff, Bruker .d, Thermo .raw, and .mzML. Additionally, Spectronaut® can process post-translational modification (PTM) DIA datasets, including phosphorylation, acetylation, and ubiquitination.
3) Acquisition: Spectronaut® can be accessed through BIOGNOSYS’s official website. A license must be purchased for long-term use.
Reference
1. Eng, J.K., A.L. McCormack, and J.R. Yates, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom, 1994. 5(11): p. 976-89.
2. Perkins, D.N., et al., Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis, 1999. 20(18): p. 3551-67.
3. Cox, J. and M. Mann, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol, 2008. 26(12): p. 1367-72.
4. Cox, J., et al., Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics, 2014. 13(9): p. 2513-26.
5. Tyanova, S., T. Temu, and J. Cox, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc, 2016. 11(12): p. 2301-2319.
6. Sinitcyn, P., et al., MaxDIA enables library-based and library-free data-independent acquisition proteomics. Nat Biotechnol, 2021. 39(12): p. 1563-1573.
7. Kong, A.T., et al., MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods, 2017. 14(5): p. 513-520.
8. Demichev, V., et al., DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods, 2020. 17(1): p. 41-44.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


