Exploring Disease Mechanisms: Key Factors in Proteomic Analysis
Proteins play a fundamental role in shaping the structure and function of tissues and organs. For example, in the progression of atherosclerosis, IL-1 promotes plaque rupture and thrombus formation, highlighting the significant impact of proteins in disease development. Consequently, proteomics has emerged as the preferred method in disease research.
To address the question of which protein-related aspects are indispensable in studies of disease mechanisms, we conducted an in-depth analysis of nearly 20 proteomic research articles published in prestigious journals such as Proteomics (IF: 5.393) and Journal of Proteome Research (IF: 5.370). Our examination encompassed various factors, including experimental design, selection of proteomic techniques, and analytical content. As a result, we identified the critical elements necessary for effective proteomic studies of diseases.
01. Importance of Sample Grouping in Disease Mechanism Studies
In research articles aimed at uncovering disease mechanisms through proteomic analysis, the grouping of samples is pivotal. The number of groups varies with no specific pattern, determined by practical needs. However, at a minimum, establishing experimental and control groups is essential. For studies involving gradient conditions, multiple groups are recommended to ensure experimental integrity. For example, when investigating the mechanism of cervical cancer regulated by lncRNA BANCR, only two groups were designated: BANCR knockdown and blank-treated HeLa cells. In studies on cancer cachexia, four groups were established: blank control, tumor without cachexia, and moderate and severe cachexia groups, aiming to differentiate cachexia severity.
|
Paper Title |
Disease |
Species |
Grouping Setup |
|
Proteomic characteristics of plasma and blood cells in natura aging rhesusmonkeys |
Aging |
Rhesus |
2 groups: old rhesus n=13; young rhesus n=13 |
|
Skeletalmuscle proteome expression differentiates severity of cancercachexia in mice and identifieslossof fragileX mental retardation syndrome-related protein 1 |
Tumor Cachexia |
Mouse |
4 groups: control n=4; tumor without cachexia n=6; moderate cachexia n=7; severe cachexia n=6 |
|
Fingolimod effects on the brain are mediatedthrough biochemica modulation of bioenergetics,autophagy,and neuroinflammatory networks |
Drug Mechanism |
Mouse |
2 groups: drug treatment n=5; control n=5 |
|
Exposing the Brain Proteomic Signatures of Alzheimer's Disease in Diverse RacialGroups: Leveraging Multiple Data Sets and Machine Learning |
Alzheimer's Disease |
Human |
2 groups:AD patients n=303;control n=196 |
|
Kaplan-Meier plots showing the effectof DKK3(top)and DKK4(bottom)expression on survival probability in the TCGA-COAD cohort.Left panels include the whole cohort middle panels show patients with mutations in SMAD4,and right panels showpatients without SMAD4 mutations |
Ovarian Cancer |
Human |
2 groups: HGSOC patients n=26; control n=24 |
|
Systematic Survey of the Regulatory Networks of the Long Noncoding RNA BANCR in Cervical CancerCells |
Cervical Cancer |
Cell |
2 groups: treatment n=3; control n=3 |
|
ProteomicStudy of the AdaptiveMechanism ofCiprofloxacin-Resistant Staphylococcus aureus totheHost Environment |
Drug Resistance |
Cell |
2 groups: treatmentA n=3; treatment B n=3 |
Biological replication is also crucial. An analysis of 20 articles reveals a pattern in biological replication based on sample types. Clinical samples generally involve a significant number, with at least 10 or more cases, sometimes even reaching hundreds. For instance, in studies on late pregnancy pre-eclampsia, 15 pre-eclampsia patient cases and samples from normal pregnant women were analyzed. In research on advanced serous ovarian cancer, 26 patient samples and 24 serum samples from healthy individuals were examined. For animal models like mice, the number of replicates is typically N≥5. For example, in studies on cancer cachexia, each group had 6 or 7 biological replicates. Investigations on fenugreek treatment mechanism involved 5 mice per drug treatment and blank control group, respectively. For cell and microbial samples, n=3 is generally sufficient for article publication, as seen in the study on cervical cancer regulation mechanism by lncRNA BANCR, with three biological replicates of BANCR knockdown cells.
02. Advances in Proteomic Techniques
Among 18 surveyed proteomic articles, except for one using PEA technology and another using SOMAscan technology, the remaining studies relied on mass spectrometry-based methods. Detailed statistical data is provided in the figure below, revealing a predominant use of TMT/iTRAQ techniques. In omics research, more data is typically considered advantageous. Compared to traditional proteomics, TMT/iTRAQ methods offer higher protein detection rates than DIA and label-free approaches. While there is typically a recommended limit on the number of samples for TMT (usually not exceeding 16), it meets the requirements for most studies on disease mechanisms, thus explaining its prevalence in this survey.
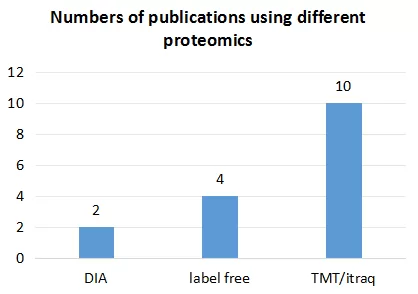
However, with the advent of 4D proteomics, there has been a shift in technological preference. Proteomic research has long pursued techniques that offer "more, faster, more accurate, and more stable" results. Traditional TMT, label-free, and DIA mass spectrometers are limited in protein detection due to their scanning speed and sensitivity. In contrast, instruments from the timsTOF Pro series, utilizing unique Tims technology and PASEF scanning mode, effectively enhance scanning speed, instrument sensitivity, identification accuracy, and detection stability, aligning more closely with the needs of proteomic research. Actual data demonstrates that 4D-DIA technology significantly outperforms traditional TMT, label-free, and DIA techniques in protein detection. Consequently, 4D proteomics technology is increasingly favored by scholars.
03. Analytical Content Presentation in Disease Proteomics
After analyzing 18 articles, we identified 13 primary components commonly presented, including: differential protein selection, PCA analysis, volcano plot, Venn diagram, cluster analysis, GO analysis, KEGG analysis, protein abundance bar chart, ROC analysis, clinical phenotype correlation analysis, protein correlation analysis, interaction network diagram, and survival analysis. Among these, the top six most frequently utilized analyses are: differential protein selection, GO analysis, cluster analysis, KEGG analysis, and partial protein abundance display. Differential protein selection serves as a fundamental aspect in articles, while GO analysis, cluster analysis, and KEGG analysis mainly focus on data mining and screening key candidate proteins. Additionally, partial protein abundance display is primarily used to illustrate and explain candidate proteins. These top six analyses are crucial in proteomic research, providing researchers with areas to focus on for data mining and enriching their articles in the field of disease proteomics research.
04. Validation Methods for Proteomic Findings
Out of the 18 articles surveyed, a remarkable 72% exhibited validation of candidate proteins identified through proteomic screening, leaving only 5 articles without validation. Various validation techniques were employed, including Western Blot, ELISA, qPCR for quantitative gene expression analysis, PRM for targeted protein detection, and immunofluorescence assay. Articles commonly utilized one or more of these methods for validation, with Western Blot being the predominant choice, followed by ELISA and qPCR.
|
Validation Methods |
Application Times |
Application Percent |
|
Western Blot |
11 |
85% |
|
ELISA |
3 |
23% |
|
qPCR |
2 |
15% |
|
PRM |
1 |
8% |
|
Immunofluorescence |
1 |
8% |
|
More than 2 methods |
4 |
31% |
MetwareBio offers cutting-edge proteomics services. If you have any needs, don't hesitate to contact us.
Read more:
· A Guide to Protein Database Selection
· MetwareBio Launches Proteomics Services
· What is Isoelectric Points of Amino Acids: Essential Calculations and Practical Applications
· Comparison and Application of Proteomic Technologies
· Demystifying Proteomics Research Strategies and Content in a Single Read
· Optimal Protein Database Selection: Insights from Experimental Data
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


