Epigenetic Modulators: How Histone Modifications Shape Gene Expression
The diversity and complexity of protein modifications grant proteins extraordinary versatility, often referred to as their “superpower of transformation.” This diversity is not only reflected in the different types of modifications but also in their reversibility and spatiotemporal specificity. These modifications influence not only protein structure and function but also participate in numerous physiological processes within cells, such as signal transduction and cell cycle regulation.
Histone modifications and histone variants play a critical role in regulating gene expression, maintaining chromatin structure, and contributing to the development of diseases. A deeper understanding of the mechanisms behind these modifications and variants can provide valuable insights into gene regulation processes and pave the way for developing new therapeutic strategies. In this blog, we will explore the regulatory mechanisms of various histone modifications and histone variants.
Histone Modifications and Chromatin Dynamics
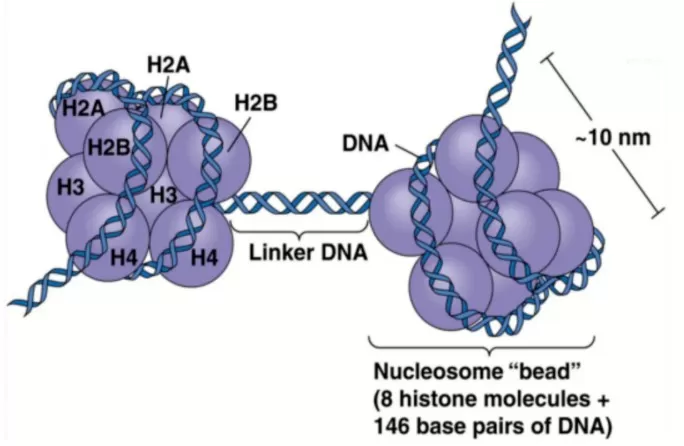 Genomic DNA is wrapped around histones to form the fundamental unit of chromatin known as the nucleosome. Each nucleosome consists of a core octamer made up of dimers of histones H2A, H2B, H3, and H4, around which approximately 147 base pairs of DNA are wound. Histone H1 acts as a linker protein, stabilizing the nucleosome structure. This arrangement provides a dynamic balance between DNA protection and transcription regulation, with flexibility stemming not only from the nucleosome structure itself but also from post-translational modifications (PTMs) of histones.
Genomic DNA is wrapped around histones to form the fundamental unit of chromatin known as the nucleosome. Each nucleosome consists of a core octamer made up of dimers of histones H2A, H2B, H3, and H4, around which approximately 147 base pairs of DNA are wound. Histone H1 acts as a linker protein, stabilizing the nucleosome structure. This arrangement provides a dynamic balance between DNA protection and transcription regulation, with flexibility stemming not only from the nucleosome structure itself but also from post-translational modifications (PTMs) of histones.
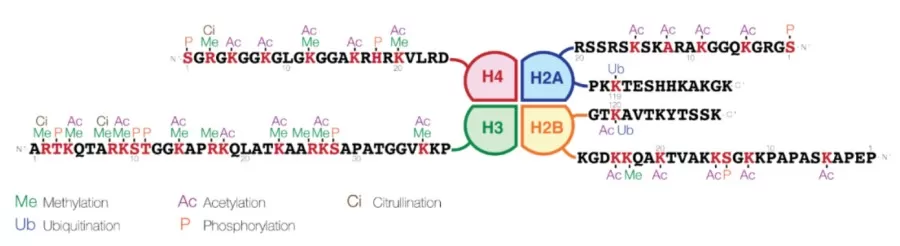
Histone modifications are a key area of epigenetic research, often referred to as the "molecular code" due to their complexity and diversity. These modifications include various chemical changes such as methylation, acetylation, phosphorylation, ubiquitination, and SUMOylation. They not only affect chromatin structure but also play a crucial role in regulating gene expression. The dynamic nature of histone modifications exemplifies this "molecular code" in action. For example, histone methylation is often associated with gene silencing, while demethylation promotes gene activation. Acetylation is typically linked to gene activation, whereas deacetylation tends to suppress gene expression. Additionally, different histone variants play critical roles in chromatin structure. For instance, certain modification sites on the histone variant H3.3 are more prominent during transcriptional activation than on the canonical H3.
Histone Methylation and Demethylation
Histone Methylation and Demethylation are key mechanisms in epigenetics that regulate chromatin structure and gene expression, influencing a wide range of biological processes.
- Histone methylation typically occurs on lysine residues of histones H3 and H4 and can involve mono-, di-, or tri-methylation. These modifications can either activate or repress gene transcription, depending on the context.
- Histone Demethylation is catalyzed by specific demethylase enzymes, which remove methyl groups from methylated lysine residues, restoring them to their unmodified state. Like methylation, demethylation is a dynamic and reversible process.
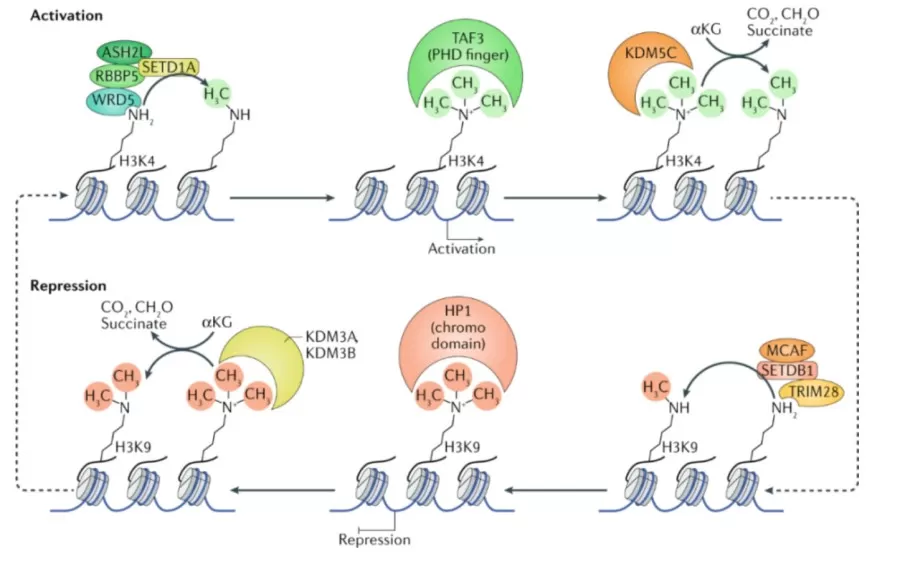
The intricate balance between histone methylation and demethylation involves numerous enzymes and regulatory mechanisms, making it a complex and highly controlled system. These modifications not only impact gene expression and chromatin structure but also play critical roles in processes such as tumorigenesis, cellular differentiation, and disease progression.
Histone Acetylation and Deacetylation
Histone acetylation and deacetylation are crucial mechanisms for regulating gene expression, involving a complex array of enzymes and molecular processes.
- Histone Acetylation is catalyzed by histone acetyltransferases (HATs), which add acetyl groups to lysine residues on histones. This reduces the positive charge of histones, decreasing their affinity for the negatively charged DNA. As a result, the chromatin structure becomes more relaxed, facilitating the binding of transcription factors and co-activators to specific DNA sequences, thereby promoting gene transcription. This process plays a pivotal role in various biological activities, including neurodevelopment, immune responses, and tumorigenesis.
- Histone Deacetylation, mediated by histone deacetylases (HDACs), removes acetyl groups, leading to a more condensed chromatin structure and repression of gene expression.
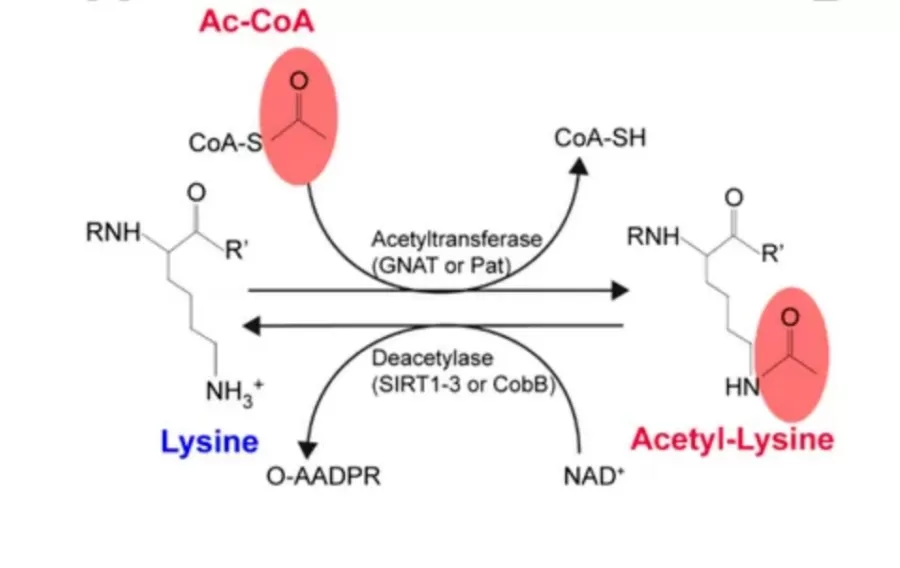
The balance between histone acetylation and deacetylation is essential for maintaining proper gene regulation. For instance, in tumor cells, histones often exhibit low acetylation levels, a condition closely linked to cancer development and progression. As a result, research into HDACs and the development of HDAC inhibitors has become a significant focus in anti-cancer drug design.
Different Types of Histone Variants
Histone variants are specialized forms of histones that differ from the canonical H2A, H2B, H3, and H4. They play key roles in regulating chromatin structure and gene expression. These variants, along with histone modifications, participate in the epigenetic regulation of chromatin and can drive the formation of distinct chromatin configurations. Common histone variants include:
1. CENPA (CenH3): This is a variant of H3 found specifically at the centromere, and it is highly conserved across eukaryotes. CENPA directs the assembly of kinetochore proteins and is crucial for chromosome segregation during cell division.
2. H3.3: With only four amino acid differences from H3, H3.3 is enriched in transcriptionally active regions, particularly in the polytene chromosomes of fruit flies, where it plays a role in transcriptional activation.
3. H2A.X: This variant is primarily involved in the DNA damage response. Its phosphorylation marks sites of DNA double-strand breaks, playing a pivotal role in repair processes like non-homologous end joining (NHEJ), homologous recombination (HR), and replication-associated DNA repair.
4. H2A.Z: An important histone variant, H2A.Z is found in regions such as the inactive X chromosome in mammals. It forms nucleosomes that are less stable than those containing canonical H2A, contributing to a more dynamic chromatin environment.
5. H2ABbd: This variant is only found on the inactive X chromosome in male rats and also forms nucleosomes that are less stable than those containing typical H2A.
6. spH2B: This histone variant is expressed in limited tissues, such as in sperm cells, and contributes to chromatin condensation during spermatogenesis.
Other histone variants, such as those incorporated by specific chromatin remodeling complexes like SWR1 and HIRA, also play essential roles in chromatin structure and function. The presence and distribution of these variants suggest that certain chromatin regions, entire chromosomes, or specific tissues may have unique "chromatin functionalities."
Reference:
1. Jiang D, Li T, Guo C, Tang TS, Liu H. Small molecule modulators of chromatin remodeling: from neurodevelopment to neurodegeneration. Cell Biosci. 2023 Jan 16;13(1):10. doi: 10.1186/s13578-023-00953-4. PMID: 36647159; PMCID: PMC9841685.
2. Shao LW, Peng Q, Dong M, Gao K, Li Y, Li Y, Li CY, Liu Y. Histone deacetylase HDA-1 modulates mitochondrial stress response and longevity. Nat Commun. 2020 Sep 15;11(1):4639. doi: 10.1038/s41467-020-18501-w. PMID: 32934238; PMCID: PMC7493924.
3. Ibarra-Morales D, Rauer M, Quarato P, Rabbani L, Zenk F, Schulte-Sasse M, Cardamone F, Gomez-Auli A, Cecere G, Iovino N. Histone variant H2A.Z regulates zygotic genome activation. Nat Commun. 2021 Dec 1;12(1):7002. doi: 10.1038/s41467-021-27125-7. PMID: 34853314; PMCID: PMC8636486.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


