Edman Degradation: A Classic Protein Sequencing Technique
Protein sequencing is a fundamental aspect of proteomics, enabling researchers to understand the structure, function, and biological significance of proteins. Among the many sequencing techniques developed over the years, Edman Degradation remains one of the most classic and precise methods for determining the N-terminal amino acid sequence of proteins. Despite the rise of high-throughput mass spectrometry (MS) techniques, Edman Degradation continues to play a crucial role in specific research applications, particularly in cases that require high-precision sequencing of short peptides. This article provides a comprehensive overview of Edman Degradation, covering its origins and development, principles, workflow, strengths and limitations, and its relevance in modern proteomics.
The Origins and Development of Edman Degradation
_1742175851_WNo_150d219.webp) The Edman Degradation method was first introduced by the Swedish scientist Pehr Victor Edman in the 1950s. Born in 1916, Edman initially pursued a career in medicine but later shifted his focus to biochemical research. Inspired by the growing interest in protein chemistry, he sought to develop a more efficient and precise technique for determining the amino acid sequences of proteins.
The Edman Degradation method was first introduced by the Swedish scientist Pehr Victor Edman in the 1950s. Born in 1916, Edman initially pursued a career in medicine but later shifted his focus to biochemical research. Inspired by the growing interest in protein chemistry, he sought to develop a more efficient and precise technique for determining the amino acid sequences of proteins.
Before Edman’s breakthrough, protein sequencing relied on methods that required complete hydrolysis of proteins into their constituent amino acids, followed by chromatographic separation and identification. These approaches provided information on amino acid composition but failed to reveal the exact sequence of residues within a peptide. To address this limitation, researchers sought a method that could determine sequences without breaking the entire protein structure apart.
In 1950, Edman proposed a stepwise sequencing technique that selectively removed only the N-terminal amino acid without disrupting the rest of the peptide. He introduced phenyl isothiocyanate (PITC) as a reagent that could label the N-terminal residue, allowing its subsequent cleavage and identification. This method marked a revolutionary shift in protein chemistry, as it enabled sequential identification of amino acids without complete protein degradation.
By the 1970s, advancements in instrumentation led to the development of automated protein sequencers, which significantly improved the efficiency of Edman Degradation. The first automated sequencing machine, developed by Edman himself, streamlined the repetitive chemical reactions required for sequencing and minimized manual errors. Later refinements, such as the incorporation of high-performance liquid chromatography (HPLC) for more precise amino acid detection, further enhanced the method’s accuracy and usability.
Throughout the 1980s and 1990s, Edman Degradation became the gold standard for sequencing small peptides, particularly in structural and functional studies of proteins. However, with the rise of mass spectrometry techniques in the late 1990s and early 2000s, the role of Edman Degradation began to diminish. Despite this, the method remains valuable in specific applications requiring high precision, such as verifying N-terminal sequences of newly discovered proteins or validating monoclonal antibodies.
The Principles of Edman Degradation
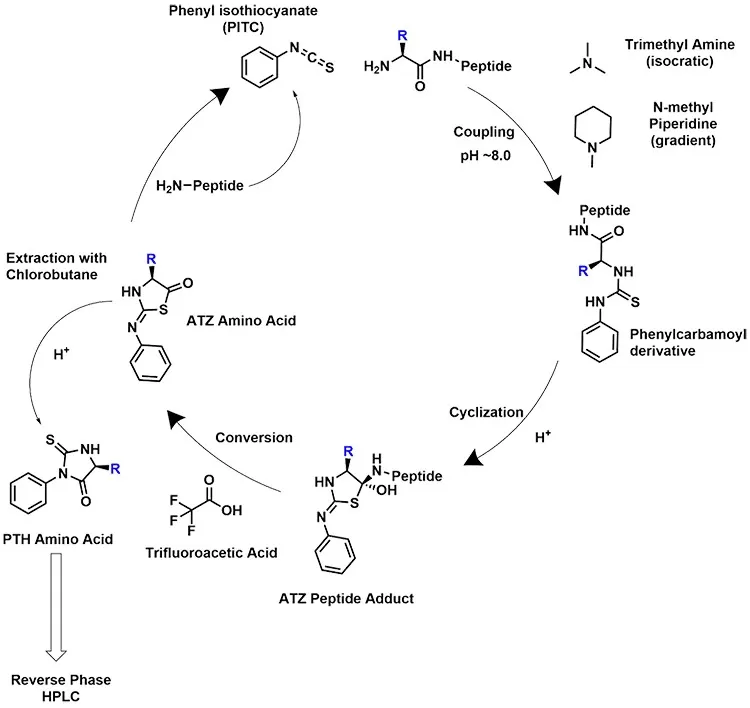
Edman Degradation is a stepwise protein sequencing method based on chemical reactions that selectively remove and identify amino acids from the N-terminus of a peptide chain. The process relies on phenyl isothiocyanate (PITC), a reagent that reacts specifically with the N-terminal amino acid, forming a stable derivative. Through controlled acid treatment, the modified amino acid is cleaved from the peptide without disrupting the rest of the sequence. This cycle is repeated, allowing the sequential identification of amino acids one by one.
Compared to earlier sequencing methods that required complete hydrolysis of proteins, Edman’s approach provided a much more precise and efficient method for sequencing. The ability to sequentially determine amino acids without completely breaking down the peptide was a major breakthrough, paving the way for detailed protein structural studies.
Work principle of Edman Degradation
Step-by-Step Workflow of Edman Degradation
The sequencing process using Edman Degradation involves multiple crucial steps, each designed to ensure the accurate removal and identification of amino acids from the peptide sequence. The method follows a cycle of chemical reactions that can be summarized as follows:
1. Coupling Reaction (Labeling the N-terminal Amino Acid)
In the first step, PITC reacts with the free amino group of the N-terminal residue under alkaline conditions, forming a phenylthiocarbamyl (PTC) derivative. This selective labeling is essential as it allows the targeted removal of one amino acid at a time without disrupting the remaining peptide chain.
2. Cyclization and Cleavage of the Labeled Amino Acid
The labeled N-terminal amino acid undergoes cyclization under acidic conditions, leading to its cleavage from the peptide chain. This reaction results in the formation of an amino acid-thiazolinone (ATZ) derivative, which is an unstable intermediate.
3. Conversion of ATZ to a Stable Derivative
The ATZ derivative is then extracted using organic solvents such as ethyl acetate and converted into a more stable form known as the phenylthiohydantoin (PTH) derivative. This transformation ensures that the released amino acid can be accurately analyzed in the next step.
4. High-Performance Liquid Chromatography (HPLC) Analysis
The PTH derivative is analyzed using high-performance liquid chromatography (HPLC) to determine the identity of the released amino acid. The separation and comparison against known standards provide precise sequence information.
5. Repeating the Process
Once the first amino acid is identified, the next residue at the N-terminus is exposed, allowing the cycle to be repeated. This iterative process continues until a complete sequence is determined or until the process is no longer efficient.
Strengths and Limitations of Edman Degradation
Edman Degradation remains a powerful tool in protein sequencing due to its precision and reliability. One of its greatest advantages is its ability to provide highly accurate sequence information, particularly for short peptides and single proteins. Since the method sequentially removes and identifies each amino acid from the N-terminus, it ensures minimal ambiguity in sequence determination, making it a preferred technique for targeted analyses. Additionally, Edman Degradation requires only small sample sizes, making it an ideal choice for analyzing specific proteins or peptide fragments that may not be easily detectable by other methods.
However, despite its strengths, Edman Degradation has notable limitations that restrict its application in all sequencing scenarios. One major challenge is its dependency on a free N-terminal amino group. If a protein’s N-terminus is chemically modified—such as through acetylation or methylation—the sequencing reaction is blocked, making the method ineffective. This limitation prevents the analysis of certain post-translationally modified proteins.
Another limitation is its inefficiency in handling long peptide chains or whole proteins. Due to cumulative material loss during repeated cycles, Edman Degradation is most effective for peptides of approximately 20-30 amino acids in length. Beyond this range, sequencing accuracy declines, and the accumulation of minor errors makes it unsuitable for large-scale proteomic studies. Additionally, some secondary structures in peptides may hinder the reaction, affecting sequencing efficiency.
Compared to mass spectrometry-based approaches, Edman Degradation is also significantly slower. While it provides high accuracy, it is a sequential process that requires multiple reaction cycles for full sequence determination. In contrast, modern mass spectrometry techniques can analyze entire protein sequences in a fraction of the time, making them more suitable for high-throughput proteomics.
The Role of Edman Degradation in Modern Proteomics
Despite the dominance of mass spectrometry in proteomics, Edman Degradation continues to be an important tool in specialized applications. It remains relevant in the validation of N-terminal sequences of newly identified proteins and in antibody research and biopharmaceutical development, where high sequencing precision is required.
Furthermore, advances in automation and integration with other analytical techniques have extended the utility of Edman Degradation. By combining it with liquid chromatography and mass spectrometry, researchers can achieve a more comprehensive analysis of protein structures, leveraging the strengths of each method for enhanced accuracy and depth.
Reference
Edman, P.; Högfeldt, Erik; Sillén, Lars Gunnar; Kinell, Per-Olof (1950). Method for determination of the amino acid sequence in peptides. Acta Chem. Scand. 4: 283–293. doi:10.3891/acta.chem.scand.04-0283.
Edman P, Begg G (1967). A protein sequenator. Eur. J. Biochem. 1 (1): 80–91. doi:10.1111/j.1432-1033.1967.tb00047.x. PMID 6059350


