DIA Proteomics vs DDA Proteomics: A Comprehensive Comparison
In the realm of mass spectrometry, the choice between Data-Independent Acquisition (DIA) and Data-Dependent Acquisition (DDA) can significantly impact the outcome of proteomic analyses. Each method offers distinct advantages, tailored to different research needs. This comprehensive comparison delves into the intricacies of DIA and DDA, exploring their unique strengths, applications, and how to choose the right technique for your specific scientific endeavors. Whether you're aiming for high sensitivity or comprehensive data coverage, understanding these techniques is crucial for advancing your research in proteomics.
1.What is DIA in Mass Spectrometry?
In the realm of mass spectrometry, the technique known as data-independent acquisition (DIA) stands out for its ability to procure and scrutinize extensive datasets with a precision and replicability that parallels the efficiency of a well-oiled machine. Unlike traditional methods, DIA fragments all analyte ions in a sample simultaneously, rather than selectively targeting specific ions based on their intensity or abundance.
The fundamental process of DIA involves the comprehensive fragmentation of each analyte ion present in the sample. Following this, the mass-to-charge (m/z) ratios of these fragments are measured. This generates a spectrum of fragment ions, known as a "mass spectrum," which can be meticulously analyzed to identify the various components within the sample. This methodical and impartial approach ensures that all potential analytes are captured within a predetermined m/z range, providing a holistic view of the sample's composition.
In contrast to data-dependent acquisition (DDA), where the instrument selects specific ions to fragment based on their abundance, DIA captures all fragment ions in a systematic manner. This unbiased method allows for the detection and quantification of every detectable analyte in the sample, regardless of their abundance or m/z value. This is particularly beneficial in complex biological samples where the presence of low-abundance analytes is critical for comprehensive analysis.
DIA is extensively used in various domains, including proteomics, metabolomics, and lipidomics. It generates exhaustive datasets that can be analyzed using advanced computational tools to extract significant biological information. This capability makes DIA a powerful technique for researchers aiming to understand the intricate details of biological systems. By leveraging DIA, scientists can gain deeper insights into the molecular underpinnings of health and disease, ultimately contributing to advancements in biomedical research.
2.What is DDA in Mass Spectrometry?
In the intricate and multifaceted world of mass spectrometry, the technique known as data-dependent acquisition (DDA) plays a pivotal role. This method is distinguished by its approach of selectively fragmenting ions based on their intensity or abundance within a sample.
Within the framework of DDA, the mass spectrometer operates by isolating a specific set of ions from the sample. These selected ions are then fragmented into smaller peptide fragments, producing a spectrum of their constituent peptides. The selection criteria for these ions are primarily based on their intensity, with the most abundant ions being prioritized for fragmentation. This targeted approach allows for detailed analysis and identification of the most prominent components within the sample.
Once the chosen ions have been effectively fragmented, the mass spectrometer proceeds to isolate and fragment the next set of ions. This process is iteratively repeated, enabling the mass spectrometer to sequentially identify and quantify a sufficient number of peptides. By focusing on the most abundant ions, DDA ensures that the resulting data is both precise and comprehensive, facilitating in-depth analysis of complex biological samples.
The DDA technique is extensively utilized in proteomics and other fields requiring detailed molecular analysis. Its ability to selectively fragment and analyze the most abundant ions makes it a valuable tool for researchers aiming to decipher the molecular intricacies of biological systems. Despite its targeted nature, DDA complements DIA by providing high-resolution data that enhances our understanding of complex biological processes.
3.Advantages of DIA
High Throughput
One of the most significant advantages of data-independent acquisition (DIA) is its ability to achieve high throughput. By fragmenting all analyte ions within a sample simultaneously, DIA enables the rapid collection of comprehensive datasets. This high-throughput capability is crucial for large-scale studies, such as proteomics and metabolomics, where the analysis of numerous samples is necessary. The efficiency of DIA allows researchers to process and analyze more samples in a shorter amount of time, accelerating the pace of scientific discovery and enabling more extensive studies.
Reproducibility
Reproducibility is another major advantage of DIA. Because DIA captures all fragment ions in an unbiased manner, the results are highly consistent across different experiments and samples. This reproducibility is essential for comparative studies, where consistent and reliable data are required to draw meaningful conclusions. With DIA, researchers can be confident that their data will be reproducible, enabling them to build on previous findings and confirm hypotheses with greater accuracy.
Comprehensive Data Analysis
DIA also excels in providing comprehensive data analysis. By capturing all detectable analytes within a sample, DIA generates extensive datasets that encompass a wide range of molecular information. This comprehensive data can be analyzed using advanced computational tools to uncover significant biological insights. The ability to detect low-abundance analytes alongside more abundant ones allows for a complete understanding of the sample's molecular composition. This holistic view is particularly valuable in complex biological systems, where minor components can have significant biological implications.
4.Advantages of DDA
High Sensitivity
One of the standout advantages of data-dependent acquisition (DDA) is its high sensitivity. By selectively fragmenting ions based on their intensity, DDA can focus on the most abundant ions within a sample. This targeted approach allows for the detection of even low-abundance peptides that might be overlooked by other methods. The high sensitivity of DDA ensures that important molecular details are captured, making it an invaluable tool for detailed proteomic analysis and the identification of novel biomarkers.
Targeted Data Collection
DDA excels in targeted data collection. By prioritizing the fragmentation of the most intense ions, DDA ensures that the resulting data is highly specific and focused on the most relevant components of the sample. This targeted approach allows researchers to concentrate their analysis on specific peptides or proteins of interest, facilitating in-depth studies of particular biological pathways or processes. The ability to selectively target and analyze specific ions makes DDA a powerful technique for hypothesis-driven research, where precise data on particular molecules is crucial.
Established Protocols
Another significant advantage of DDA is the availability of established protocols. Over the years, extensive research and development have led to the optimization of DDA methods, resulting in well-established protocols that are widely accepted in the scientific community. These protocols provide a reliable framework for conducting DDA experiments, ensuring consistency and reproducibility across different studies. Researchers can leverage these established protocols to streamline their experimental design and data analysis, reducing the time and effort required to achieve accurate and meaningful results. This consistency is particularly valuable in comparative studies and large-scale projects where reproducibility is paramount.
5.Why is DIA Better than DDA?
When it comes to analyzing complex proteomic samples, data-independent acquisition (DIA) stands out as the superior method over data-dependent acquisition (DDA) for several compelling reasons. The advantages of DIA include its ability to detect low-abundance peptides, its increased specificity, and its greater reproducibility. These factors collectively make DIA an indispensable tool for researchers in drug discovery, clinical research, and beyond, facilitating deeper insights into complex biological systems and diseases.
Detection of Low-Abundance Peptides
One of the most significant limitations of DDA is its propensity to generate incomplete or biased data. This is primarily due to its potential to miss low-abundance peptides and its bias towards highly abundant ones. DDA selectively fragments ions based on their intensity, which can lead to the omission of less abundant but potentially critical peptides. In contrast, DIA fragments all ions within a pre-defined m/z range, ensuring comprehensive coverage and enabling the detection of low-abundance peptides that DDA might overlook. This comprehensive fragmentation approach allows DIA to provide a more complete and accurate representation of the sample's proteome.
Increased Specificity
Another advantage of DIA over DDA is its increased specificity. DIA can differentiate between isobaric peptides, which have the same m/z but different sequences, by simultaneously fragmenting multiple precursor ions and using their fragment ions to distinguish between them. This results in a more precise and accurate analysis of complex proteomic samples. The ability to accurately differentiate and identify peptides even in the presence of isobaric interferences makes DIA a powerful tool for detailed proteomic studies where high specificity is essential.
Greater Reproducibility
Reproducibility is a critical factor in proteomics, especially in fields such as drug discovery and clinical research, where consistent and reliable results are imperative. DIA boasts greater reproducibility compared to DDA because it analyzes all ions within a pre-defined m/z range in every run. This ensures consistent coverage and accuracy across multiple samples, reducing the variability that can arise from selective ion fragmentation in DDA. By providing a more reliable and consistent method for analyzing complex proteomic samples, DIA enhances the confidence in the results and their applicability to real-world biological questions.
6.How to Choose DIA and DDA Quantitative Proteomics
In the captivating world of proteomics, selecting the right mass spectrometry technique is essential for obtaining accurate and reliable results. Data-Independent Acquisition (DIA) and Data-Dependent Acquisition (DDA) are two of the most widely used techniques. However, making the right choice between these methods depends on several critical factors that can significantly influence the outcome of your study.
Research Goal
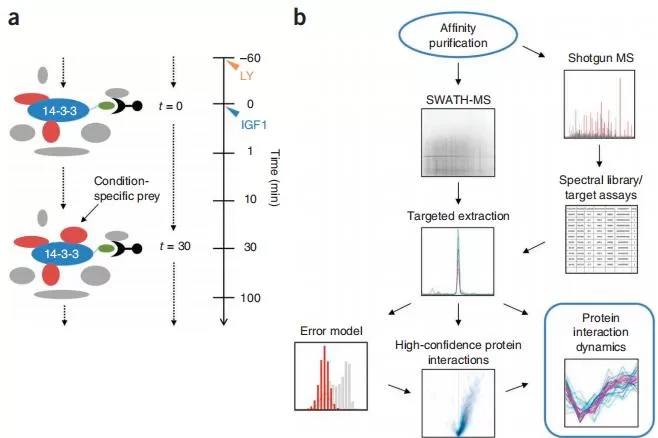
Your research objectives play a pivotal role in determining whether to use DIA or DDA. DIA is renowned for its high coverage and quantitative accuracy, making it ideal for large-scale protein studies. For instance, Collins et al. (2017) effectively used DIA to quantify over 9,000 proteins in the mouse brain, showcasing its capability to analyze extensive proteomes.
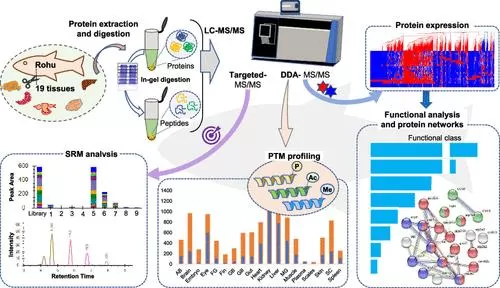
Conversely, DDA excels in small-scale studies requiring high sensitivity and precision, particularly in detecting post-translational modifications. A study by Mehar Un Nissa et al. (2022) utilized DDA to analyze the proteome and post-translational modifications of tropical water fish, highlighting DDA's sensitivity for such applications.
Sample Type
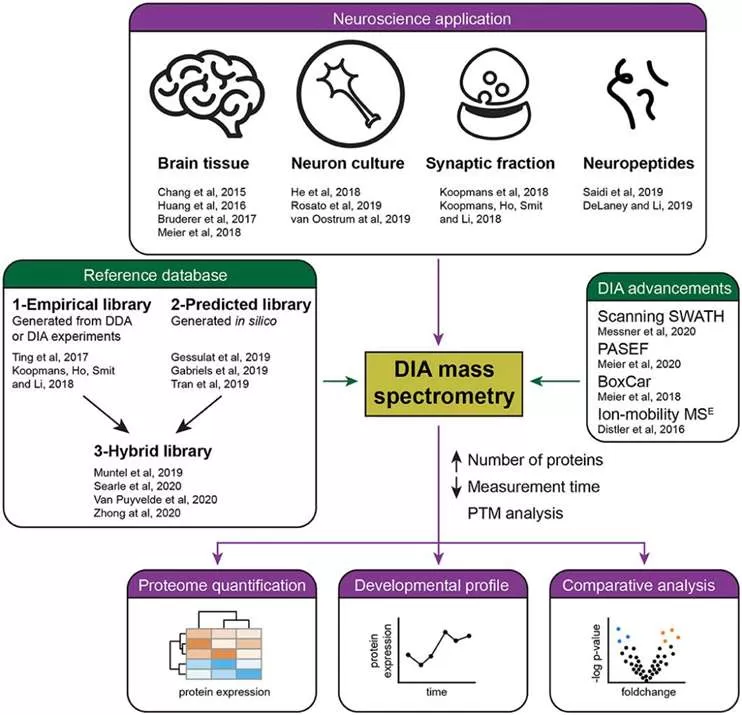
The complexity of your sample also dictates the choice between DIA and DDA. DIA is optimal for analyzing complex samples like whole cells or tissues. Ka Wan Li et al. (2020) demonstrated the efficacy of DIA in brain research, particularly in quantitative analyses of cellular and synaptic proteomes, revealing the spatial and temporal changes in proteins related to neuronal plasticity and disease mechanisms. On the other hand, DDA is better suited for less complex samples, such as purified protein extracts, offering high sensitivity and accuracy in simpler biological contexts.
Data Analysis
Data complexity and analysis requirements are crucial considerations. DIA generates highly complex data necessitating advanced bioinformatics tools for analysis. I. Ortea et al. (2016) used various bioinformatics tools to analyze the proteome of human bronchoalveolar lavage fluid, illustrating the need for sophisticated software to handle DIA's data complexity and achieve comprehensive proteome analysis.
In contrast, DDA produces simpler data that can be analyzed with more straightforward approaches. While this simplicity makes DDA data easier to interpret using less sophisticated tools, it may limit the depth of biological insights due to less comprehensive proteome coverage.
Conclusion: Making the Right Choice for Your Proteomics Research
In conclusion, both DIA and DDA offer unique advantages that cater to different research requirements in proteomics. While DDA provides high sensitivity and targeted data collection, DIA ensures comprehensive data coverage and reproducibility. The choice between these methods should be guided by your specific research objectives, sample complexity, and resource availability.
For researchers seeking expert guidance and advanced proteomics services, MetwareBio is a leading multiomics CRO provider in the field. Explore our DIA Proteomics, DDA Proteomics, Serum/Plasma Proteomics, and Phosphoproteomics services to elevate your research and achieve accurate, reliable results.
FAQ
Q1: What is the difference between DIA and DDA proteomics?
Data-Dependent Acquisition (DDA) selects high-intensity peptide ions for fragmentation in mass spectrometry, offering high sensitivity but missing low-abundance peptides and reducing reproducibility. Data-Independent Acquisition (DIA), like SWATH-MS, fragments all peptides in predefined m/z windows, ensuring comprehensive proteome profiling and better consistency. DDA suits targeted studies; DIA excels in discovery-driven research but requires advanced bioinformatics.
Q2: What are the advantages of DIA over DDA?
DIA proteomics outperforms DDA by offering comprehensive peptide coverage, capturing low-abundance proteins, and improving reproducibility across runs. It enhances quantitative proteomics for post-translational modifications (PTMs) and supports retrospective analysis with robust datasets. Ideal for biomarker discovery in complex samples, DIA’s systematic approach minimizes missing data.
Q3: What is the difference between DIA and Ida?
Information-Dependent Acquisition (IDA), synonymous with DDA, targets high-intensity peptides for fragmentation in mass spectrometry, prioritizing sensitivity but risking inconsistent sampling. Data-Independent Acquisition (DIA) fragments all peptides in m/z windows, offering unbiased, reproducible proteome analysis. IDA suits targeted proteomics; DIA excels in discovery, requiring advanced computational tools. Metware tailors both approaches for precise protein profiling.
Q4: What are the three types of proteomics?
The three main types of proteomics are Bottom-Up Proteomics, which digests proteins into peptides for LC-MS analysis, ideal for high-throughput proteome profiling; Top-Down Proteomics, analyzing intact proteins to study proteoforms and PTMs; and Targeted Proteomics, using SRM or PRM for precise quantification of specific proteins.
References:
-
Collins, B., Gillet, L., Rosenberger, G. et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat Methods 2013.
-
Mehar Un Nissa et al. Organ-Based Proteome and Post-Translational Modification Profiling of a Widely Cultivated Tropical Water Fish, Labeo rohita Journal of Proteome Research 2022
-
Ortea, A. et al. Discovery of potential protein biomarkers of lung adenocarcinoma in bronchoalveolar lavage fluid by SWATH MS data-independent acquisition and targeted data extraction, Journal of Proteomics 2016
-
Ka Wan Li et al (2020). Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome Front. Mol. Neurosci., 23 December 2020
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.