How Evolutionary Blueprints Are Paving the Way for Nutrient-Rich CoQ10 Crops
We are thrilled to highlight a groundbreaking study titled "Design of CoQ10 Crops Based on Evolutionary History" published in Cell (2025). This interdisciplinary research combined evolutionary biology, widely-targeted metabolomics, and advanced genome editing to decode the genetic basis of coenzyme Q (CoQ) side-chain variations in plants. By analyzing CoQ profiles across 134 plant species and leveraging plant metabolomics data, the team identified key amino acid residues controlling CoQ chain length. Their findings enabled the engineering of CoQ10-producing rice and wheat—a leap toward addressing global nutritional gaps. MetwareBio’s proprietary metabolomics platform played a pivotal role in quantifying CoQ isoforms, demonstrating the power of multi-omics in crop biofortification.
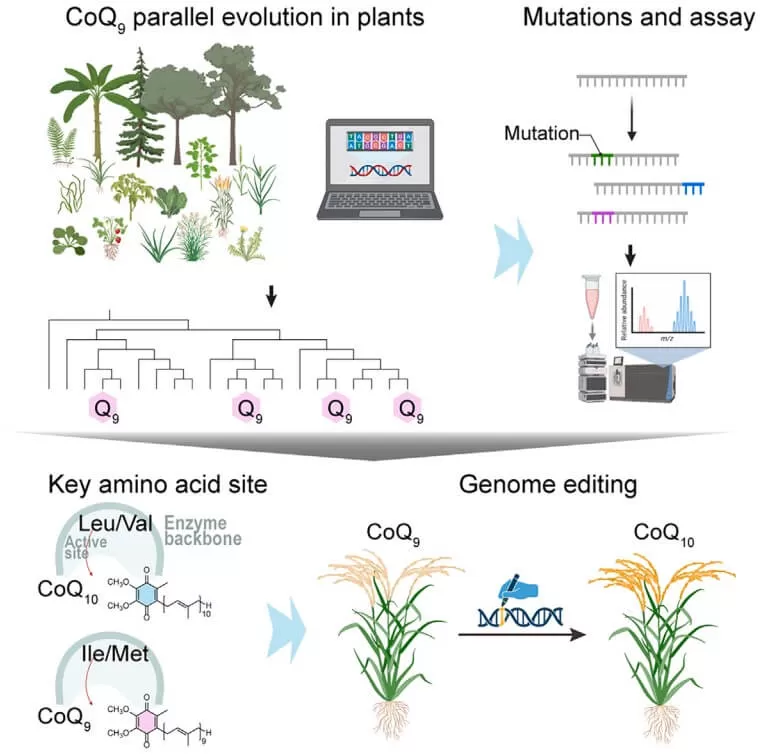
Design of CoQ10 crops based on evolutionary history
The Critical Role of CoQ10 in Human Health
Coenzyme Q10 (CoQ10) is a vital antioxidant and mitochondrial electron carrier, essential for cardiovascular health and cellular energy production. While humans synthesize CoQ10, dietary intake remains crucial, especially for aging populations or individuals on statin therapy. Unfortunately, staple crops like rice and wheat predominantly produce CoQ9, a shorter-chain variant with limited bioavailability. Enhancing CoQ10 levels in crops could transform global nutrition, but this goal has long been hindered by unclear molecular mechanisms. This study bridges that gap by tracing CoQ evolution across land plants and deploying cutting-edge metabolomic profiling and genome editing to redesign crops.
CoQ10 as the Ancestral Trait in Flowering Plants
Using LC-MS/MS-based metabolomics, researchers mapped CoQ9 and CoQ10 distribution across 134 plant species, including ferns, gymnosperms, and angiosperms. They discovered that CoQ10 production is ancestral in flowering plants, while CoQ9 emerged independently in herbaceous lineages like Poaceae (grasses) and Cucurbitaceae. This evolutionary divergence was linked to mutations in the Coq1 gene, which encodes the enzyme responsible for polyisoprenoid chain elongation.
 of Co_1739857988_WNo_1244d681.webp)
A time-calibrated molecular phylogeny of 134 land plants with marginal ancestral state reconstruction (ASR) of CoQ10 or CoQ9 states
Site 240: The Genetic Switch for Chain Length
Through comparative genomics and machine learning-driven feature selection, the team pinpointed amino acid residue 240 in Coq1 as the critical determinant of chain length. Substitutions at this site (e.g., leucine to methionine) repeatedly drove CoQ9 evolution. Structural modeling and molecular dynamics simulations revealed how residue 240 stabilizes intermediate products during chain elongation, with hydrophobic interactions dictating final chain length.
Prime Editing Unlocks CoQ10 in Staple Crops
Guided by evolutionary insights, researchers used multiplex prime editing to modify native Coq1 genes in rice and wheat. Introducing five key amino acid changes in rice shifted CoQ9 production to CoQ10, achieving >75% CoQ10 in grains without compromising yield. Similar edits in wheat also showed significant CoQ10 accumulation, marking a milestone in metabolic engineering.
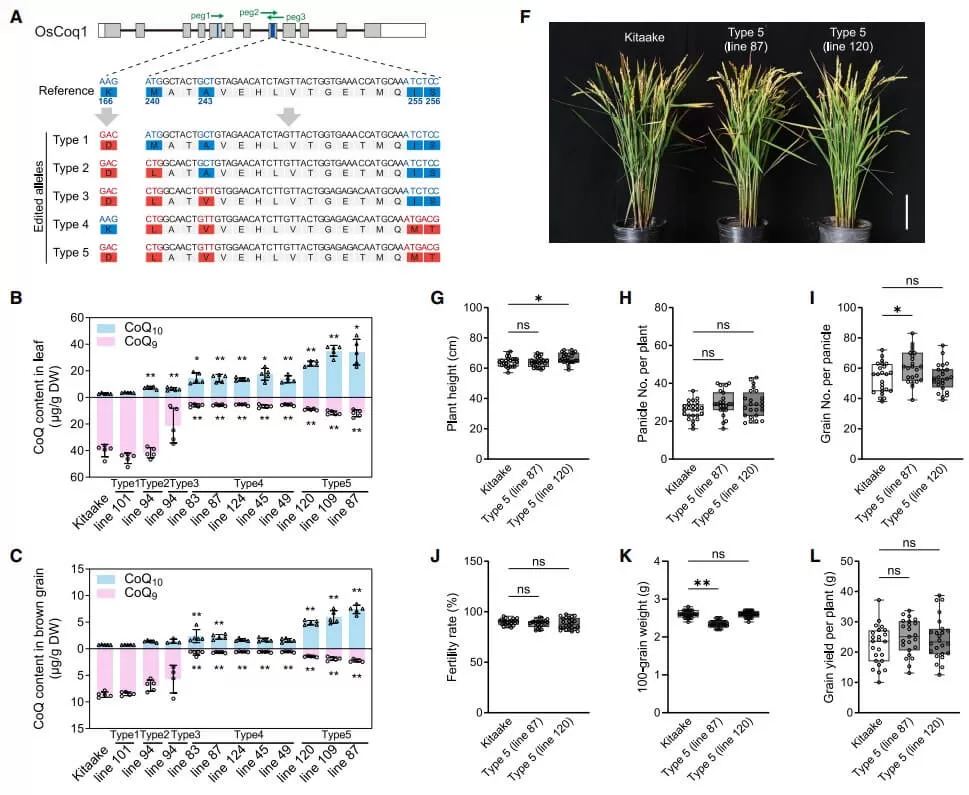
Generating CoQ10 rice lines by genome editing
Multi-Omics as the Catalyst for Nutritional Innovation
This study exemplifies how integrating evolutionary metabolomics, genomics, and genome editing can solve longstanding agricultural challenges. By reconstructing ancestral CoQ states and identifying conserved molecular mechanisms, the team transformed a fundamental biological question into a practical solution for human health. The use of widely-targeted metabolomics was indispensable for quantifying CoQ isoforms across diverse species, underscoring its value in comparative biology. Meanwhile, prime editing’s precision highlights its potential for crop improvement, avoiding transgenic controversies. Looking ahead, this approach could be extended to other nutrients (e.g., vitamins, antioxidants), positioning multi-omics as the cornerstone of next-generation biofortification.
Partner with MetwareBio for Transformative Crop Studies
At MetwareBio, we specialize in leveraging cutting-edge metabolomics to drive transformative advancements in plant research. Our widely-targeted metabolomics, powered by high-resolution LC-MS/MS systems, enables precise quantification of over 3,000 metabolites, offering unparalleled insights into plant metabolic pathways. With our newly expanded in-house plant metabolite database, now encompassing 60,000 metabolites, we are uniquely equipped to support diverse plant studies, from crop biofortification to stress response analysis.
Partner with us to unlock the full potential of metabolomics in your plant studies and achieve groundbreaking results.
Reference:
Xu JJ, Lei Y, Zhang XF, Li JX, Lin Q, Wu XD, Jiang YG, Zhang W, Qian R, Xiong SY, Tan K, Jia Y, Zhou Q, Jiang Y, Fan H, Huang YB, Wang LJ, Liu JY, Kong Y, Zhao Q, Yang L, Liu J, Hu YH, Zhan S, Gao C, Chen XY. Design of CoQ10 crops based on evolutionary history. Cell. 2025 Feb 11:S0092-8674(25)00087-X. doi: 10.1016/j.cell.2025.01.023.


