Decoding ICP: Proteomics Insights into Biomarkers and Pathways
Unraveling Intrahepatic Cholestasis of Pregnancy: Role of lncRNAs
Intrahepatic cholestasis of pregnancy (ICP) is a liver disorder that occurs specifically during pregnancy. Characterized by intense itching (pruritus) without an associated rash and sometimes accompanied by jaundice, ICP typically manifests in the third trimester and can lead to several complications for both the mother and the fetus. The exact cause of ICP is not fully understood, but it is thought to involve genetic, hormonal, and environmental factors. Elevated bile acids in the blood, due to impaired bile flow from the liver, are a hallmark of the condition. ICP is associated with poor perinatal outcomes, including fetal distress, preterm birth, and, in severe cases, unexpected intrauterine death. Despite its serious implications, the mechanisms by which ICP leads to these adverse outcomes remain unclear, making it a critical area of research in maternal-fetal medicine.
Long noncoding RNAs (lncRNAs), which are transcripts over 200 nucleotides long, are increasingly recognized for their roles in various biological processes, including cancer, inflammation, and cell autophagy. Recent studies have highlighted their involvement in intrahepatic cholestasis of pregnancy (ICP). Notable lncRNAs such as ENST00000505175.1, ASO3480, and ENST00000449605.1 have shown potential as diagnostic markers for ICP based on serum levels. Additionally, Linc02527 is found to be elevated in ICP patients' placenta and serum, promoting autophagy and trophoblast cell proliferation, which may contribute to ICP pathology. However, the exact roles of lncRNAs in placental function and fetal development in ICP remain largely unexplored. While current research suggests that lncRNAs could be crucial in regulatory networks affecting disease progression, further investigation is needed to clarify their functions and interactions. Understanding these mechanisms could lead to noninvasive diagnostic tools and new therapeutic targets for managing ICP.
Proteomics: Unlocking the Mysteries of the Protein Universe
Proteomics is the large-scale study of proteins, which are vital components of living organisms and play crucial roles in biological processes. Unlike genomics, which focuses on the study of genes, proteomics deals with the entire set of proteins expressed by a cell, tissue, or organism at a certain time. This field is essential for understanding the structure, function, and interactions of proteins, which can provide insights into cellular processes, disease mechanisms, and potential therapeutic targets.
Data-Independent Acquisition (DIA) Quantitative Proteomics is a cutting-edge technique in proteomics that allows for comprehensive and reproducible analysis of proteins in complex biological samples. Unlike traditional methods, which may selectively analyze only a subset of proteins, DIA captures data for all detectable proteins in a sample in a single run. This approach improves the coverage and quantification accuracy of the proteome, making it especially valuable in identifying and quantifying proteins across different samples or conditions. DIA proteomics is widely used in research to discover biomarkers, study disease mechanisms, and understand biological pathways at a detailed level.
Proteomic Profiling of ICP: Unraveling Autophagy and Regulatory Networks
To better understand the mechanisms of ICP and develop effective treatments or preventative strategies, a research team published an article on the journal Frontiers in Cell and Developmental Biology titled “Comprehensive Analysis of Quantitative Proteomics with DIA Mass Spectrometry and ceRNA Network in Intrahepatic Cholestasis of Pregnancy” (article resource). The study reveals that core proteins—SPI1, FOXK1, SLC13A3, and MBD2, along with their upstream lncRNAs and miRNAs, may be potential drug targets for alleviating adverse perinatal outcomes in ICP patients.
The authors firstly constructed the rat ICP model and collected placentas from 10 normal pregnancies and 10 ICP patients. Then they conducted DIA quantitative proteomics analyses to the placentas and GO, KEGG, COG/KOG functional enrichment analyses to the proteomics data. The results found 77 upregulated proteins and 99 downregulated proteins and the differentially expressed proteins were implicated in a wide range of biological processes, such as the process utilizing the autophagic mechanism, autophagy, protein acetylation, porphyrin-containing compound metabolic process, and pigment metabolic process.
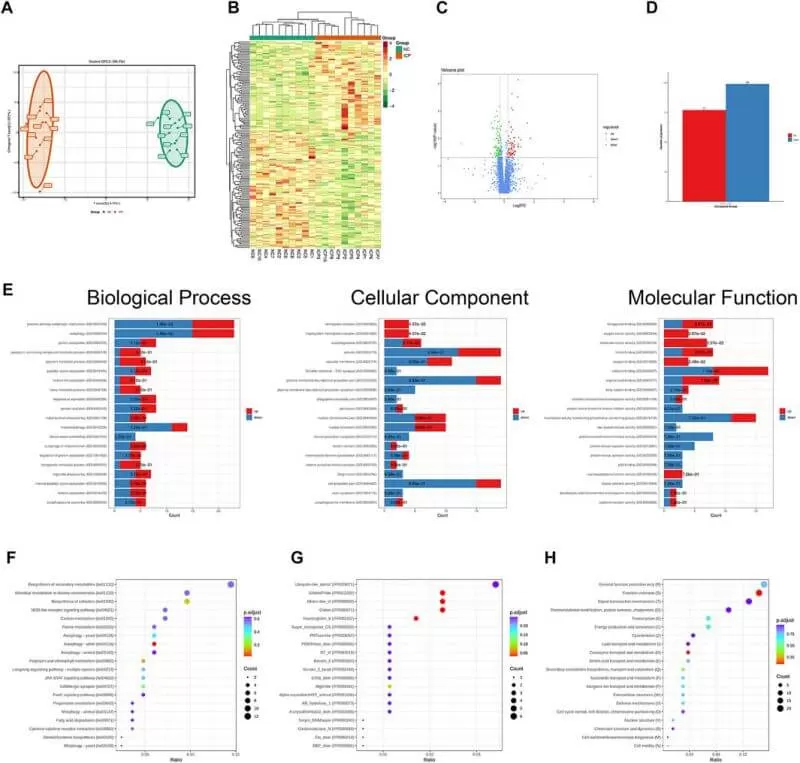
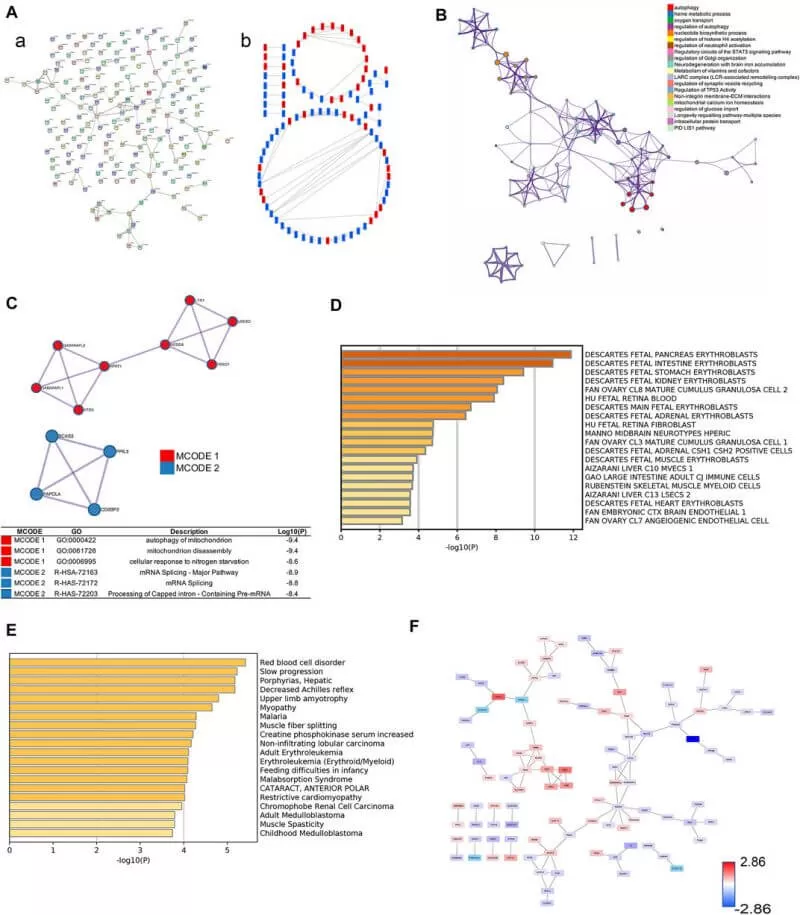
Then the authors selected core proteins from the prediction of the protein–protein interaction network using STRING, Cytoscape and Metascape databases. The proteins in the PPI network were ranked by the degree algorithm, and the results showed top 20 proteins were NEDD8, ATG5, FECH, ACAT2, HBE1, ACOX3, CTPS2, GABARAPL2, GABARAPL1, MLST8, PLEK, RANBP6, HMBS, PRKAG2, SDC1, PBDC1, ECI2, ACE, PEX14, and GNG10. The MCODE algorithm was applied to identify densely connected network components and showed that FBXO7, LTN1, UBE2O, NEDD8, HPRT1, GANARAPL1, GABARAPL2, and ATG5 were included in MCODE1, and CD2BP2, PPIL3, PAPOLA, and BCS2 were components of MCODE2. Furthermore, the proteins in MCODE 1 were associated with autophagy of the mitochondrion, mitochondrion disassembly, and cellular response to nitrogen starvation. The proteins in MCODE 2 were connected with the mRNA splicing-major pathway, mRNA splicing, and processing of capped intron-containing pre-mRNA. The results of enrichment analyses in DisGeNET and PaGenBase showed that ICP development was closely associated with delivery gestational age, and HBG1, SPI1, HBG2, HBE1, FOXK1, KRT72, SLC13A3, MBD2, SP9, GPLD1, MYH7, and BLOC1S1 were linked to the occurrence of ICP.
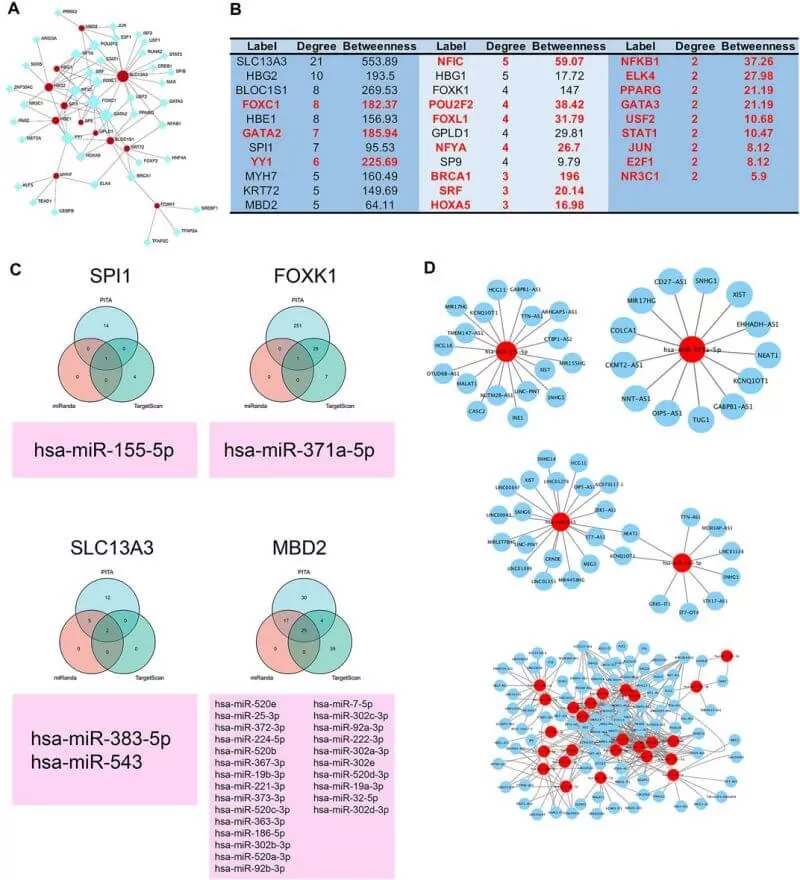
Finally, to explore the regulatory mechanism, the authors constructed the transcription factor-protein network and found a variety of miRNAs and lncRNAs participated in the regulation of core proteins expression. And the authors conducted validation to their results using in vitro and in vivo experimental models. The Western blotting results of the explant demonstrated that compared with the NC group, the expressions of autophagy-related proteins—Beclin 1 and LC3 II/1 were reduced significantly, which was consistent with their quantitative proteomics.
MetwareBio: Your Trusted Partner in Proteomics Solutions
MetwareBio had offered the DIA quantitative proteomics service for this research. MetwareBio is a multiomics CRO focusing on developing and applying innovative multiomics technologies to life science and health research. With a dedicated commitment to data quality and a nuanced understanding of the unique nature of each project, MetwareBio offers tailored metabolomics, proteomics and multi-omics combination analyses services to suit diverse needs. Whether it's small-scale endeavors or large population studies, our workflows are adept at accommodating varying sample sizes and project scopes. Our extensive experience, reflected in over 20,000 completed projects, underscores our proficiency in delivering reliable results. At MetwareBio, we prioritize collaboration, guiding researchers from sample extraction to data analysis to ensure their research goals are met with precision and efficiency. Please don't hesitate to reach out if you have any requirements or inquiries!
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


