DDA vs. DIA: The Essential Guide to Label-Free Quantitative Proteomics
What is Label-Free Quantitative Proteomics?
Label-free quantitative proteomics (LFQ) is a technique that enables the quantification of proteins in biological samples without the need for labeling reagents. This method relies on the analysis of peptide ion intensities obtained from mass spectrometry (MS), allowing for the comparison of protein abundance across different conditions or treatments. Label-free quantitative proteomics has gained prominence in contemporary proteomics due to its simplicity and broad applicability, making it widely utilized in medical and life sciences research.
Label-free quantitative proteomics can be classified into two techniques based on the mass spectrometry acquisition modes: DDA quantitative proteomics using data-dependent acquisition mode and DIA quantitative proteomics using data-independent acquisition. Historically, DDA was referred to simply as label-free quantitative proteomics to distinguish it from labeled techniques like TMT proteomics. However, with the advent of DIA technology, which also operates without labels, we now categorize both methods under the umbrella of label-free quantitative proteomics.
How to Conduct Label-Free Quantitative Proteomics
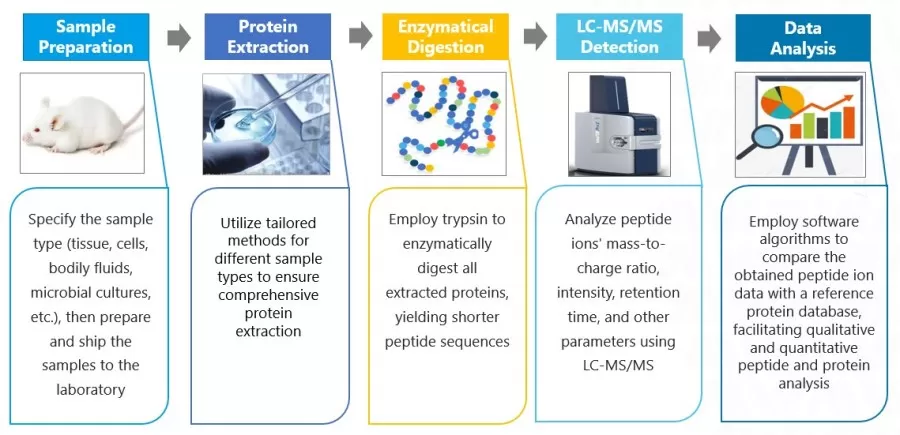
The basic workflow of label-free quantitative proteomics involves the extraction of total proteins from the sample, followed by enzymatic digestion using trypsin to generate peptides. These peptides are then subjected to mass spectrometry analysis, where qualitative and quantitative assessments are performed based on the secondary spectra and response intensities of the peptides.
DDA Quantitative Proteomics
DDA quantitative proteomics (previously known as label-free quantitative proteomics) employs a data-dependent acquisition (DDA) mode to obtain mass spectrometry data. The process begins with a full MS1 scan, during which peptide precursor ion information is recorded based on their intensities. Then, a number of precursor ions with intensity ranking highest are selected to generate their MS2. During the first MS2 scan, a narrow mass window is set around the most intense precursor ion, allowing the quadrupole to selectively filter the target peptide ions. These ions are then sent to the collision room for fragmentation, and the resulting fragment ions are recorded in terms of their mass-to-charge ratios and intensities. This process continues with subsequent MS2 scans until a complete cycle is achieved. This approach does not rely on expensive labeling reagents, and generally involve less complex data analysis. However, the DDA mode may lose some low-abundance ion information, which can slightly reduce protein coverage depth.
DIA Quantitative Proteomics
DIA quantitative proteomics is based on a data-independent acquisition (DIA) mode to obtain mass spectrometry data. The process begins with an MS1 full scan, during which precursor ion information for peptides is recorded. Subsequently, the mass range is divided into several wide mass windows based on the mass-to-charge ratios of the precursor ions, allowing for MS2 acquisition. During each MS2 scan, specific peptide segments within the predetermined wide mass windows are filtered using the quadrupole and then sent to the collision chamber for fragmentation. The resulting fragment ions are recorded in terms of their mass-to-charge ratios and intensities. This process continues with additional MS2 scans until a complete cycle is achieved. Compared to DDA techniques, DIA captures MS2 spectra for all precursor ions, enhancing both protein coverage depth and quantitative stability.
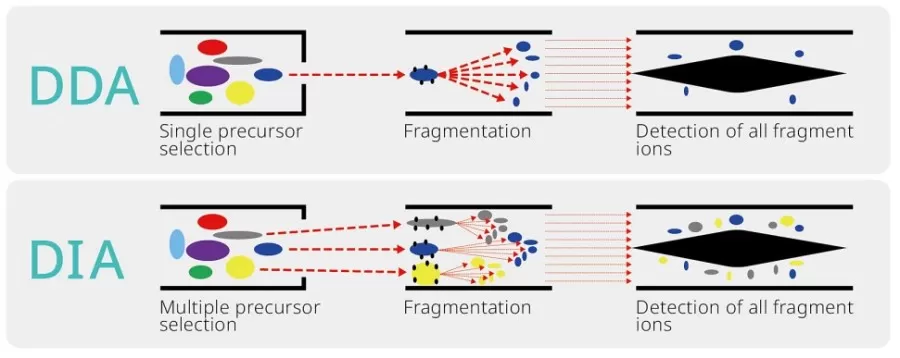
DDA vs. DIA
As the primary methods in label-free quantitative proteomics, DDA (Data-Dependent Acquisition) and DIA (Data-Independent Acquisition) each possess unique strengths and limitations that distinguish them from one another. DDA excels in its ability to select the most abundant ions for detailed analysis, resulting in high-quality spectral data. However, this can lead to issues with reproducibility and coverage in complex samples. Conversely, DIA captures data from all ions simultaneously, providing a more comprehensive view of the proteome, but it often requires advanced data analysis techniques to interpret the results effectively. In this comparison, we explore the key features, advantages, and challenges associated with both DDA and DIA proteomics, helping researchers determine the best approach for their specific applications.
|
Technique |
||
|
Scanning Mode |
DDA |
DIA |
|
Identification Level |
MS2 |
MS2 |
|
Quantification Level |
MS1 |
MS2 |
|
Advantage |
Simpler experimental process and easier data analysis |
Better protein coverage and higher reproducibility |
|
Limitation |
Lower protein coverage and higher missing values |
More complex data processing with higher analysis difficulty |
Advantages and Limitations of Label-Free Quantitative Proteomics
In the rapidly evolving field of proteomics, researchers often face the challenge of choosing between label-free and labeled techniques. Each approach presents distinct advantages and limitations that can significantly influence research outcomes. Label-free proteomics, celebrated for its simplicity and high throughput, allows for the analysis of complex biological samples without the need for intricate labeling processes. Conversely, labeled proteomics offers enhanced sensitivity and reproducibility, making it a powerful tool for precise quantification. In the section below, we clearly illustrate the advantages and limitations of label-free proteomics in comparison to labeled proteomics, providing insights to help guide researchers in selecting the most suitable method for their specific objectives.
Advantages of Label-Free Quantitative Proteomics
- Simplicity: No need for complex labeling processes, making it easier to implement and requiring less sample preparation.
- Dynamic Range: Can detect a broader range of protein concentrations, which is especially useful for analyzing complex samples.
- Quantification Flexibility: Allows for relative quantification across different conditions without the need for standards.
- High Throughput: Suitable for large-cohort studies, enabling the analysis of multiple samples simultaneously.
- Low Cost: Generally more cost-effective since it avoids the expense of labeling reagents and their associated protocols.
Limitations of Label-Free Quantitative Proteomics
- Reproducibility: Often less reproducible than labeled methods due to variations in sample processing and instrument sensitivity.
- Sensitivity: Generally lower sensitivity, making it challenging to detect low-abundance proteins compared to labeled approaches.
- Data Complexity: Data analysis can be more complex, requiring advanced algorithms to handle variability and noise.
- Quantification Variability: Relative quantification may vary significantly across runs, complicating data interpretation.
- Bias Toward Abundant Proteins: Often favors the detection of high-abundance proteins, potentially overlooking low-abundance targets.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


