Building a Cutting-Edge 4D Proteomics Platform
In recent years, with continuous advancements in technology, proteomics has moved from methodological research into practical applications, making a significant impact across various research fields, including precision medicine. However, compared to more mature genomic and transcriptomic sequencing technologies, proteomics still lags in terms of throughput, sensitivity, and quantitative reliability. These gaps primarily stem from the high complexity of the proteome, combined with the limited separation and detection capabilities of mass spectrometers. The scan speed and sensitivity of the instruments restrict the detection of proteins. For instance, in a single DDA detection experiment, a total of 101,726 signals were detected, but only 17% of these signals were used to collect secondary spectra, and just 9.6% of the signals were successfully identified. To overcome the challenges of proteomics, 4D proteomics technology has emerged.
Introduction to 4D Proteomics
Mass spectrometry-based proteomics research is fundamentally based on analyzing the physicochemical properties of sample ions, yielding qualitative and quantitative results from the sample's mass spectra and related data. 4D proteomics expands upon the traditional three-dimensional separations of retention time, m/z (mass-to-charge ratio), and ion intensity, adding a fourth dimension—collision cross-section (CCS) in ion mobility—to redefine the standards of proteomics analysis.
Advantages of 4D proteomics
- Enhanced scanning speed: >120 Hz MS/MS (the fastest scanning speed among mass spectrometers for actual sample analysis).
- Increased sensitivity: Boosted by 20 times compared to other proteomic instruments.
- Improved specificity: Ion mobility separation enhances spectrum reliability.
- Enhanced stability: Requires no internal instrument cleaning for 4 months, facilitating high-throughput sample analysis.
- Real-time database retrieval engine: Synchronizes data collection and retrieval, saving on data analysis time.
The heightened sensitivity and scanning speed of 4D proteomics substantially elevate the quality, quantity, and throughput of protein identification.
Decoding 4D Proteomics Data
1. Assured detection with minimal sample volumes (high sensitivity)
By conducting three-needle technique replicates across varying protein sample volumes, the data reveals a consistent rise in detected proteins as sample amounts increase from 1 ng to 200 ng. Notably, protein detection peaks at 6498 proteins when the sample volume reaches 200 ng.
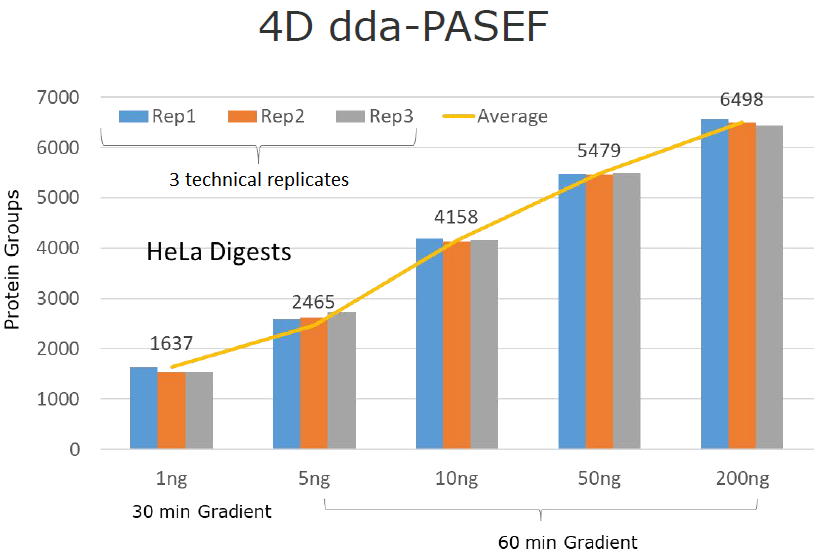 Traditional proteomic experiments typically demand microgram-level sample volumes. However, 4D proteomics is adaptable to trace samples. For instance, in exosome research, a mere 2 ml of plasma or 5 ml of whole blood suffices. This is particularly advantageous in studies involving animal models like mice, where obtaining sufficient blood samples is challenging. With 4D proteomics, the minimum required protein amount can plummet to 200 ng, just a fraction of the conventional proteomic sample, promising significant advancements in exosome proteome analysis.
Traditional proteomic experiments typically demand microgram-level sample volumes. However, 4D proteomics is adaptable to trace samples. For instance, in exosome research, a mere 2 ml of plasma or 5 ml of whole blood suffices. This is particularly advantageous in studies involving animal models like mice, where obtaining sufficient blood samples is challenging. With 4D proteomics, the minimum required protein amount can plummet to 200 ng, just a fraction of the conventional proteomic sample, promising significant advancements in exosome proteome analysis.
2. Streamlining experimental timelines while guaranteeing high-throughput sample analysis
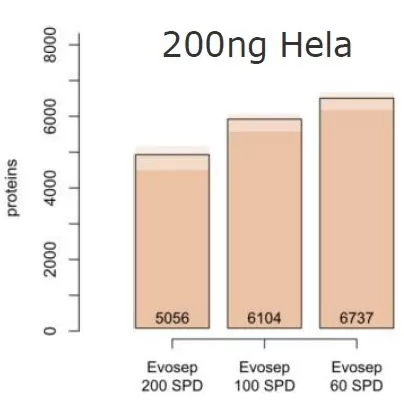
4D proteomics utilizes efficient, high-throughput chromatographic separation techniques, enabling experiments with increased throughput without sacrificing depth of protein coverage. For instance, based on actual data, with an injection volume of 200 ng of HeLa cell lysate, and a throughput of 200 samples per day, the results demonstrate the identification of 5056 proteins, effectively ensuring comprehensive protein coverage. Compared to typical proteomic experiments handling about 20 samples per day, 4D proteomics can boost throughput by tenfold, significantly reducing experimental timelines, ensuring stable quantification, and enhancing the precision of experimental outcomes, particularly suitable for large-scale sample analyses.
3. Enhanced Qualitative Precision
In the introduction to 4D proteomics, it's been highlighted for its utilization of a four-dimensional protein separation mode. This approach effectively simplifies the complexity of protein data, thus refining the accuracy of protein identification. Particularly in intricate biological samples where numerous peptides co-elute, distinguishing among them within a small retention time window can be challenging due to their minute mass differences (m/z). However, ion mobility can discern peptides with minimal mass discrepancies (distinct ion mobility), facilitating more precise identification.
For instance, consider the identification of protein modifications; when the same modification occurs at different sites, the variations in mass-to-charge ratios are negligible, making it challenging for mass spectrometry to differentiate, especially amidst co-elution during the chromatographic phase. With 4D proteomics, such concerns are alleviated. It can effectively differentiate between various modified protein sites using ion mobility, effortlessly identifying tens of thousands of modified peptide sequences, outperforming traditional high-resolution proteomic instruments by 50-100%.
4. Improved Quantitative Precision
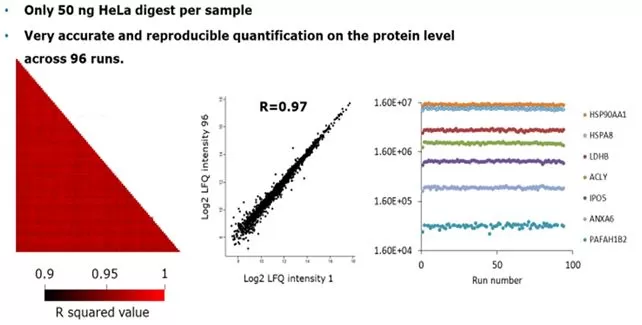
The superior stability of 4D proteomics instruments contributes to heightened quantitative accuracy. Testing a series of 96 consecutive samples reveals an average correlation coefficient of 0.95 between samples 1-96, while samples 1 and 96 exhibit an impressive correlation coefficient of 0.99. Furthermore, scrutiny of seven randomly selected peptides confirms consistent response patterns, as depicted in accompanying images. Collectively, these findings underscore the heightened stability of 4D proteomics testing, yielding more precise quantification.
Conclusion: The Future of Proteomic Research with 4D Proteomics
In summary, the 4D proteomics technology not only improves acquisition speed and sensitivity but also enables deep protein coverage even under short gradients, showcasing remarkable stability in high-throughput sample analysis. MetwareBio utilizes the timsTOF HT mass spectrometer on the 4D proteomics platform to offer enhanced services for proteomic research.
Read more:
· A Guide to Protein Database Selection
· MetwareBio Launches Proteomics Services
· What is Isoelectric Points of Amino Acids: Essential Calculations and Practical Applications
· Comparison and Application of Proteomic Technologies
· Demystifying Proteomics Research Strategies and Content in a Single Read
· Optimal Protein Database Selection: Insights from Experimental Data
· Protein sample preparation tips: Serum or Plasma?
· An Overview of Mainstream Proteomics Techniques
· Exploring Disease Mechanisms: Key Factors in Proteomic Analysis
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.


