Handling Missing Values and Outliers in Bioinformatics
In bioinformatics research, data quality directly determines the reliability and accuracy of analytical results. However, missing values and outliers are common challenges in real-world datasets. If not properly addressed, these issues can introduce significant bias into downstream analyses. Therefore, mastering appropriate techniques for handling missing values and outliers is an indispensable skill in bioinformatics.
Handling Missing Values
1. Causes of Missing Values
Missing values arise from diverse sources, and their origins vary across omics disciplines:
- Transcriptomics: Incomplete RNA extraction, low reverse transcription efficiency, insufficient sequencing depth, or data filtering during processing.
- Proteomics: Non-expression of proteins, abundance below instrumental detection limits, sample loss during preparation, peptide miscutting during digestion, low ionization efficiency, or poor peptide-spectrum matching.
- Metabolomics: Metabolite concentrations below detection thresholds, sample loss during preparation or analysis, instrumental instability, or limitations in data processing algorithms.
- Genomics/Epigenomics (supplementary): Sequencing errors, low coverage regions, or biases in amplification and alignment.
2. Methods for Handling Missing Values
(1) Deletion Methods
The simplest approach involves filtering data based on the proportion of missing values in samples or groups. For example, metabolites missing in >50% of samples may be excluded. However, deleting features with minor missingness risks losing biologically significant information. Thus, imputation methods are preferred for datasets with low missing rates.
(2) Imputation Methods
Fixed-Value Imputation: Replace missing values with constants (e.g., 0, minimum, ½ minimum, or ⅕ minimum) or statistical measures (mean, median). While straightforward, this method may introduce bias, especially when missingness is non-random.
Advanced Imputation Techniques:
- k-Nearest Neighbors (KNN): Estimates missing values using the mean of the ‘k’ most similar samples. KNN is flexible but computationally intensive and sensitive to noise. Optimal ‘k’ and distance metrics (e.g., Euclidean, Manhattan) should be selected based on data characteristics.
- Random Forest (RF): Predicts missing values by training models on observed data. RF handles non-linear relationships and complex structures but requires substantial computational resources. Hyperparameter tuning (e.g., tree depth, number of trees) is critical for performance.
- Singular Value Decomposition (SVD): Reconstructs data matrices by retaining dominant singular values. SVD reduces dimensionality and preserves key patterns but is sensitive to feature selection and computationally expensive for large datasets.
(3) Model-Based Imputation:
- Multiple Imputation by Chained Equations (MICE): Iteratively imputes missing values using regression models for each variable, accounting for uncertainty.
- Deep Learning Approaches: Autoencoders or generative adversarial networks (GANs) learn latent representations to predict missing values, particularly effective for high-dimensional omics data.
Handling Outliers
Outliers (anomalous values) are data points that deviate significantly from other observations. They may arise from experimental errors, data entry mistakes, or genuine biological variation. Proper outlier management is essential for robust statistical inference.
1. Outlier Detection Methods
(1) Box Plot
A box plot visualizes data distribution using quartiles:
- Lower Quartile (Q1), Median (Q2), Upper Quartile (Q3).
- Interquartile Range (IQR): IQR = Q3 – Q1.
- Bounds: Upper = Q3 + 1.5×IQR; Lower = Q1 – 1.5×IQR.
Values beyond these bounds are classified as outliers. Box plots are robust to outliers and ideal for exploratory data analysis.
(2) Z-Score Normalization
The Z-score (i.e., the difference from the mean divided by the standard deviation for that data point) was calculated for each data point, and points with an absolute value of the Z-score greater than 3 were usually considered outliers. For normally distributed data, compute the Z-score:
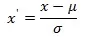
(3) Isolation Forest
This tree-based algorithm isolates outliers by recursively partitioning data. Outliers require fewer splits to be isolated, making the method efficient for large datasets. It operates without prior assumptions about data distribution.
(4) Additional Methods
- DBSCAN Clustering: Identifies outliers as points in low-density regions.
- Local Outlier Factor (LOF): Measures local deviation relative to neighbors, effective for detecting context-dependent anomalies.
2. Strategies for Managing Outliers
(1) Deletion
Remove outliers if they result from identifiable errors (e.g., measurement artifacts) and constitute a small fraction of the dataset.
(2) Correction
Replace outliers with plausible values, such as adjacent measurements, Winsorized limits, or model-based predictions (e.g., regression imputation).
(3) Retention
Retain outliers if they reflect true biological variation (e.g., rare disease subtypes) or carry critical information. Sensitivity analyses should then assess their impact on conclusions.
Summary
Handling missing values and outliers is a critical yet nuanced step in bioinformatics workflows. Key considerations include:
1. Data Context: Align methods with the biological rationale behind missingness or anomalies (e.g., technical vs. biological causes).
2. Method Trade-offs: Balance computational efficiency, scalability, and assumptions (e.g., normality for Z-scores).
3. Validation: Use cross-validation or resampling to evaluate imputation accuracy. For outliers, employ domain knowledge to verify findings.
By integrating these strategies, researchers enhance data quality, mitigate analytical biases, and lay a robust foundation for biomarker discovery and mechanistic studies. Future advancements in machine learning and multi-omics integration will further refine these approaches, enabling more precise and reproducible analyses.
Read more
Understanding WGCNA Analysis in Publications
Deciphering PCA: Unveiling Multivariate Insights in Omics Data Analysis
Metabolomic Analyses: Comparison of PCA, PLS-DA and OPLS-DA
WGCNA Explained: Everything You Need to Know
Harnessing the Power of WGCNA Analysis in Multi-Omics Data
Beginner for KEGG Pathway Analysis: The Complete Guide
GSEA Enrichment Analysis: A Quick Guide to Understanding and Applying Gene Set Enrichment Analysis
Comparative Analysis of Venn Diagrams and UpSetR in Omics Data Visualization


