Applications of Proteomics in Agriculture – Disease Resistance
In recent years, proteomics has emerged as a transformative tool in agricultural research, particularly in enhancing disease resistance in crops. As the world faces growing challenges related to food security and climate change, the ability to safeguard crops from diseases has become more crucial than ever. Proteomics, the study of proteins and their functions, offers a powerful means to unlock the molecular mechanisms behind disease resistance, providing farmers with more effective tools to combat pathogens and improve crop yields. In this article, we’ll explore how proteomics is reshaping the way we understand and manage disease resistance in agriculture, and how this innovative approach can help ensure a sustainable future for global food production.
Table of Contents
-
How Proteomics Helps in Identifying Disease Resistance Mechanisms
-
Key Techniques in Proteomics for Disease Resistance Research
-
Case Studies: Proteomics Applications in Disease Resistance of Major Crops
-
Benefits of Proteomics in Agriculture for Disease Resistance
-
Challenges in Implementing Proteomics for Disease Resistance in Agriculture
-
Future Prospects: The Role of Proteomics in Sustainable Agriculture
What is Proteomics and Why is it Important in Agriculture?
Proteomics refers to the large-scale study of proteins, particularly with regard to their functions and interactions. In agriculture, proteomics is used to analyze the protein profiles of plants to understand how they respond to diseases and environmental stress. By studying these proteins, researchers can identify key biomarkers that indicate resistance to specific pathogens, offering new ways to protect crops from diseases.
Plants, like all living organisms, produce a variety of proteins that are involved in various biological processes. Some of these proteins are directly related to disease resistance, and proteomics helps identify these proteins and understand their roles. By uncovering the molecular pathways involved in resistance, proteomics provides insights that can be used to develop more resilient crops.
How Proteomics Helps in Identifying Disease Resistance Mechanisms
Analyzing Plant Protein Profiles to Identify Disease Resistance Markers
A key aspect of proteomics in agriculture is the ability to analyze plant proteins and identify markers that are linked to disease resistance. Researchers can compare the protein profiles of resistant and susceptible plant varieties to pinpoint proteins that play a role in resistance. These markers can then be used in breeding programs to develop crops with improved resistance to diseases.
Role of Proteomics in Understanding Host-Pathogen Interactions
Another critical application of proteomics is in the study of host-pathogen interactions. When a plant is infected by a pathogen, it produces specific proteins as part of its defense mechanism. By examining these proteins, scientists can gain insights into how the plant detects the pathogen, activates its immune response, and mounts an effective defense. Understanding these interactions is essential for developing crops that can better withstand diseases.
Proteomics refers to the large-scale study of proteins, particularly with regard to their functions and interactions. In agriculture, proteomics is used to analyze the protein profiles of plants to understand how they respond to diseases and environmental stress. By studying these proteins, researchers can identify key biomarkers that indicate resistance to specific pathogens, offering new ways to protect crops from diseases. Plants, like all living organisms, produce a variety of proteins that are involved in various biological processes. Some of these proteins are directly related to disease resistance, and proteomics helps identify these proteins and understand their roles. By uncovering the molecular pathways involved in resistance, proteomics provides insights that can be used to develop more resilient crops.
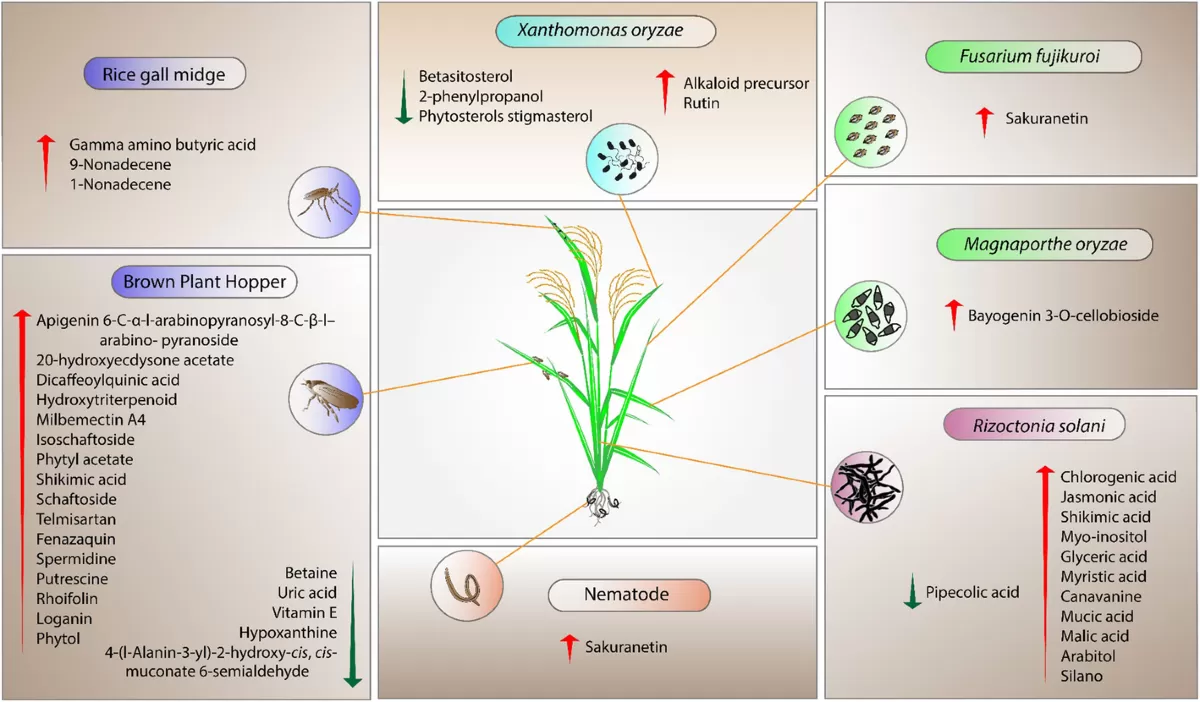
Proteomics and Metabolomics Studies on the Biotic Stress Responses of Rice (Vo et al., 2021)
Key Techniques in Proteomics for Disease Resistance Research
Mass Spectrometry in Identifying Protein Biomarkers
Mass spectrometry is one of the most powerful tools used in proteomics to identify and quantify proteins in plant tissues. This technique allows researchers to detect specific biomarkers associated with disease resistance, helping them to track the plant’s immune response to infection. Mass spectrometry also provides detailed information on protein structure, which is crucial for understanding how these proteins function in the plant’s defense system.
Two-Dimensional Gel Electrophoresis for Disease Resistance Proteins
Two-dimensional gel electrophoresis (2D-GE) is another widely used technique in proteomics. It separates proteins based on their size and charge, allowing researchers to identify and analyze a large number of proteins in a single experiment. This method is particularly useful for identifying proteins involved in disease resistance, as it provides a detailed protein map that can reveal changes in protein expression during infection.
Other Techniques for Protein Quantification and Characterization
In addition to mass spectrometry and 2D-GE, there are other techniques that are used in proteomics to study plant proteins, such as Western blotting, enzyme-linked immunosorbent assay (ELISA), and protein microarrays. These methods are used to quantify specific proteins, characterize their functions, and study their interactions with other molecules. Together, these techniques provide a comprehensive understanding of plant immunity and disease resistance.
Case Studies: Proteomics Applications in Disease Resistance of Major Crops
Rice: Proteomics in the Fight Against Rice Blast Disease
Rice blast disease, caused by the fungus Magnaporthe oryzae, is one of the most destructive diseases in rice cultivation. By using proteomics, researchers have identified proteins involved in rice’s defense response to this pathogen. These proteins, such as pathogenesis-related (PR) proteins, play a key role in the plant’s ability to recognize and defend against the pathogen. Through proteomics, scientists have developed new strategies for breeding rice varieties with enhanced resistance to blast disease, helping to protect global rice production.
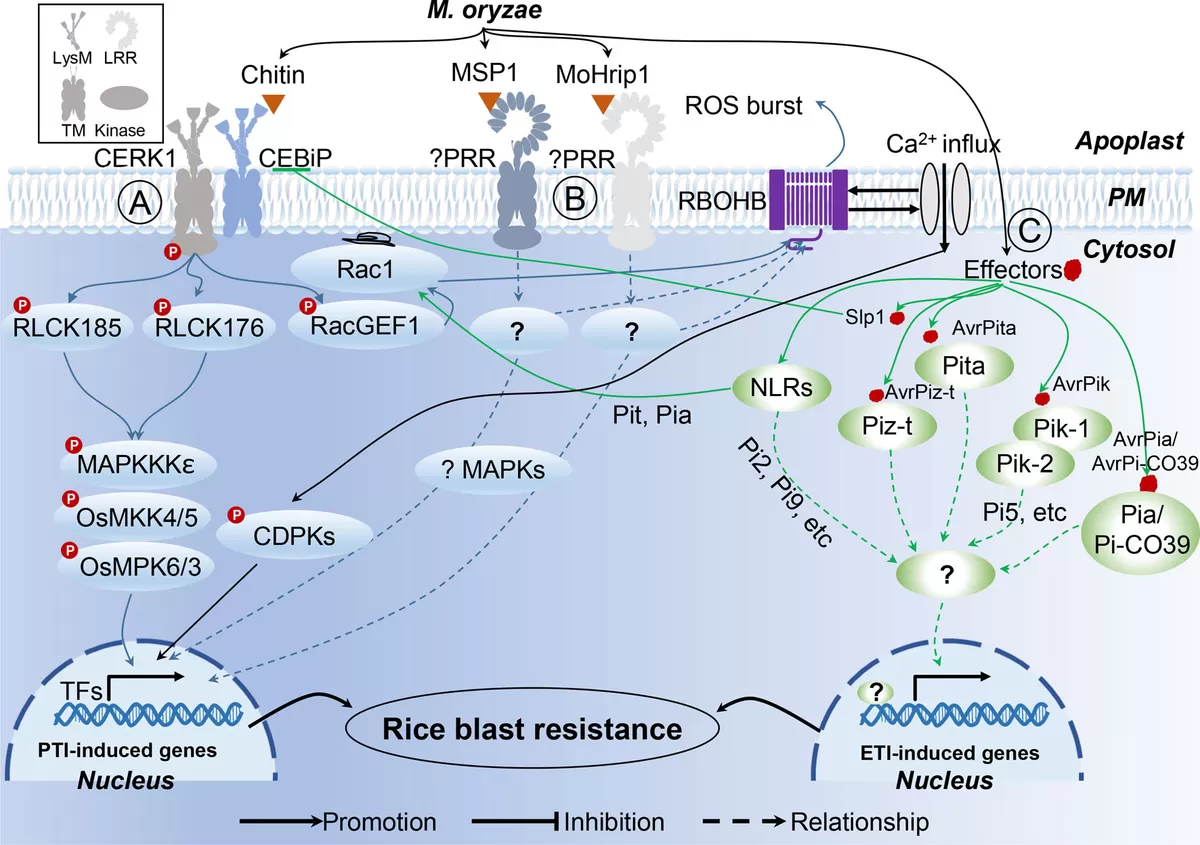
Proteomics of Rice—Magnaporthe oryzae Interaction (Meng et al., 2019)
Wheat: Identifying Disease Resistance Proteins for Wheat Stem Rust
Wheat stem rust, caused by the fungus Puccinia graminis, has historically been one of the most devastating diseases in wheat crops. Proteomics has helped identify key proteins involved in wheat’s defense against stem rust, including proteins that activate the plant’s immune response. By identifying these proteins, researchers can breed wheat varieties that are more resistant to stem rust, improving food security in wheat-growing regions.
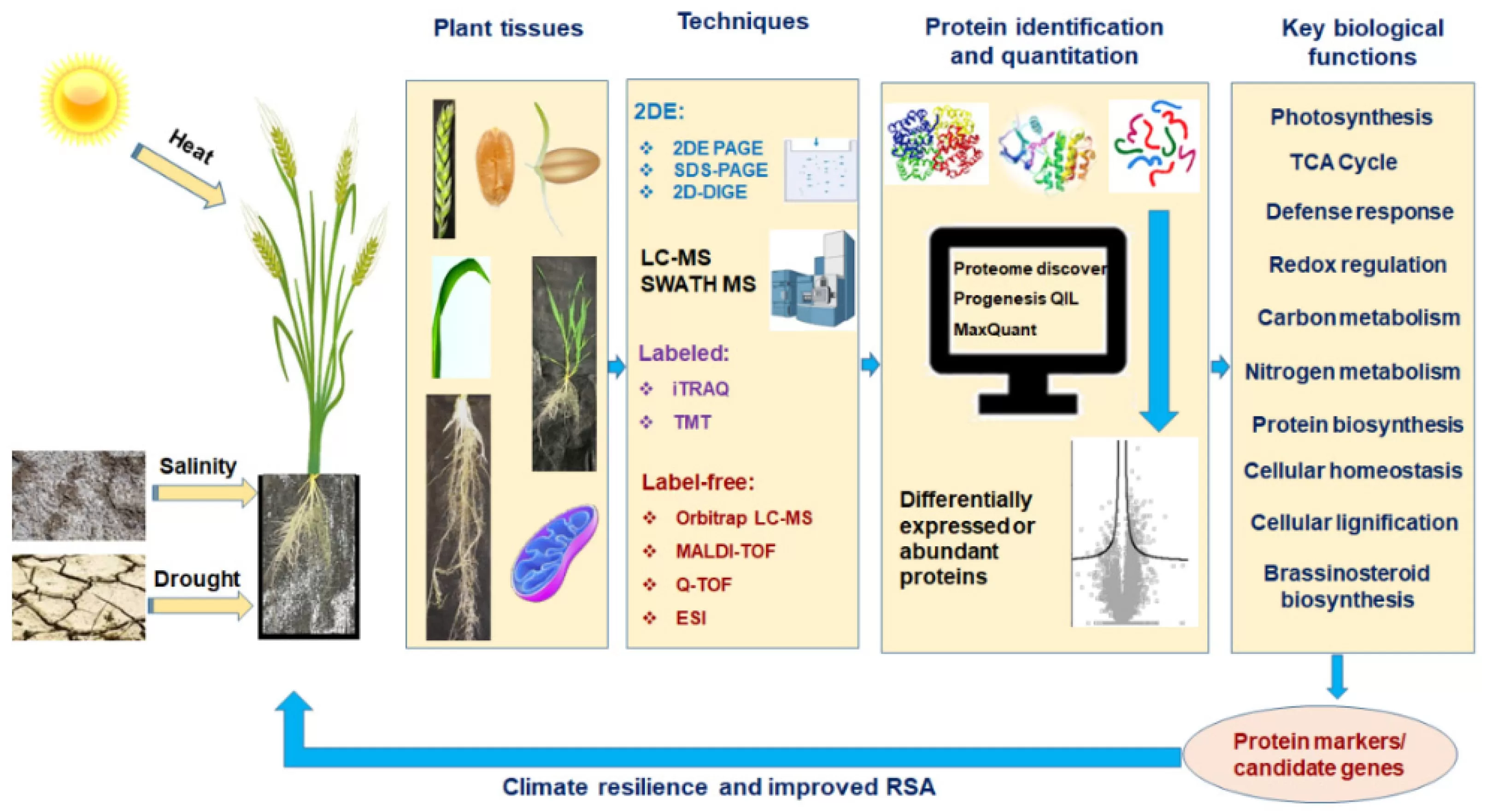
Proteomics identifying Disease Resistance Proteins for Wheat Stem Rust (Halder et al., 2022)
Tomatoes: Using Proteomics to Combat Tomato Late Blight
Late blight, caused by the oomycete Phytophthora infestans, is a major threat to tomato production. Through proteomics, researchers have identified several tomato proteins that are involved in the plant’s defense against late blight. These proteins, such as defensins and chitinases, help the plant fight off the pathogen by breaking down its cell walls. By understanding the role of these proteins, scientists are developing tomato varieties with improved resistance to late blight, reducing the need for chemical pesticides.
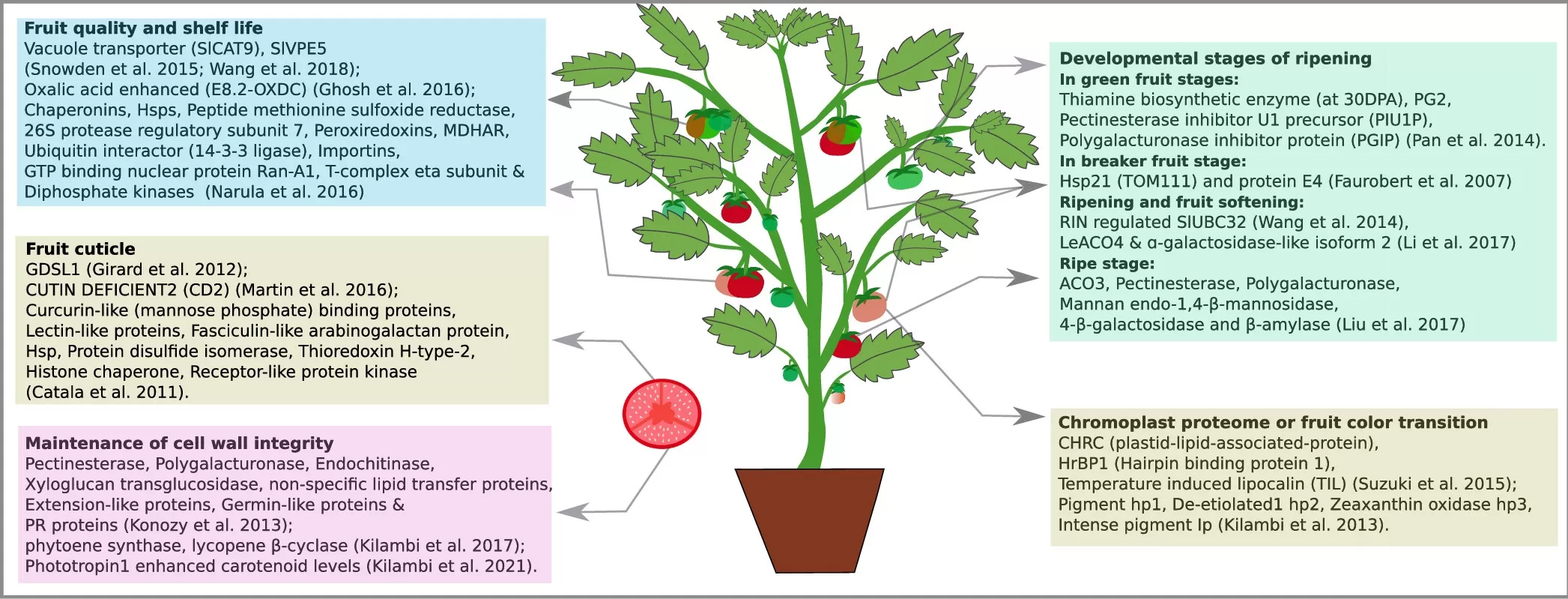
Proteomics of Reproductive Development, Fruit Ripening, and Stress Responses in Tomato (John et al., 2022)
Benefits of Proteomics in Agriculture for Disease Resistance
Faster Identification of Resistance Traits
One of the key advantages of proteomics is its ability to speed up the identification of disease resistance traits in plants. Traditional breeding methods can take years to develop resistant varieties, but proteomics allows researchers to identify proteins associated with resistance more quickly, accelerating the breeding process.
Improving Crop Yield and Reducing Dependency on Pesticides
By developing disease-resistant crops through proteomics, farmers can reduce their reliance on chemical pesticides. This not only lowers production costs but also helps protect the environment and improve the sustainability of agriculture. Disease-resistant crops are also more likely to produce higher yields, contributing to global food security.
Enhancing Crop Breeding Programs for Disease Resistance
Proteomics has revolutionized crop breeding programs by providing a more efficient way to select plants with desirable traits. By identifying the proteins involved in disease resistance, breeders can select plants that are more likely to withstand disease outbreaks, leading to healthier, more resilient crops.
Challenges in Implementing Proteomics for Disease Resistance in Agriculture
High Costs and Technical Complexity
While proteomics offers great promise for enhancing disease resistance, the technology can be expensive and technically challenging. Mass spectrometry, for example, requires specialized equipment and expertise, which can make it difficult for small-scale farmers or researchers with limited resources to access these tools.
Data Interpretation and Integration Challenges
Another challenge in proteomics research is the complexity of the data generated. Interpreting large-scale protein data requires sophisticated bioinformatics tools and expertise, which can be a barrier to widespread implementation. Additionally, integrating proteomics data with other types of molecular data, such as genomics, adds another layer of complexity.
Scaling Proteomics Applications Across Large Farms
Applying proteomics at a large scale is another challenge. While the technology has proven effective in laboratory settings and small-scale trials, scaling it up for use on large farms can be difficult. There is a need for more affordable, user-friendly tools that can be used by farmers to monitor disease resistance in real time.
The Role of Proteomics in Sustainable Agriculture
Precision Agriculture and the Role of Proteomics in Disease Management
The future of proteomics in agriculture lies in its integration with precision agriculture technologies. By combining proteomics with sensors, drones, and data analytics, farmers will be able to monitor the health of their crops in real time and make data-driven decisions about disease management. This will lead to more efficient use of resources and reduced environmental impact.
Integration with Genomics and Bioinformatics for Improved Crop Resilience
As proteomics continues to evolve, its integration with genomics and bioinformatics will provide even deeper insights into plant disease resistance. By combining data from multiple omics technologies, researchers will be able to identify new resistance mechanisms and develop crops that are even more resilient to disease and environmental stress.
Proteomics is playing a pivotal role in transforming agriculture, especially when it comes to disease resistance. With its ability to identify key proteins involved in plant immunity, proteomics is helping scientists and farmers develop more resilient crops, reduce pesticide use, and improve global food security.
While challenges remain in terms of cost and data complexity, the future of proteomics in agriculture looks promising. By embracing these advancements, we can create a more sustainable and disease-free future for agriculture.
Contact Metwarebio stay updated on the latest proteomics advancements and their potential to revolutionize disease resistance in agriculture.
FAQs
What is the difference between Proteomics and Genomics in agriculture?
Proteomics focuses on the study of proteins, while genomics focuses on the study of genes. In agriculture, both are used to improve crop traits, but proteomics provides insights into how genes are expressed and how proteins function in response to disease.
How does Proteomics contribute to the development of disease-resistant crops?
Proteomics helps identify proteins that are involved in disease resistance, which can then be used to develop crops with enhanced immunity to pathogens.
What are the challenges faced when applying Proteomics in agriculture?
Challenges include the high cost and complexity of the technology, difficulties in data interpretation, and the need to scale proteomics applications for large-scale farming.
Reference
1. Vo, K.T.X., Rahman, M.M., Rahman, M.M. et al. Proteomics and Metabolomics Studies on the Biotic Stress Responses of Rice: an Update. Rice 14, 30 (2021). https://doi.org/10.1186/s12284-021-00461-4
2. Meng Q, Gupta R, Min CW, Kwon SW, Wang Y, Je BI, Kim Y-J, Jeon J-S, Agrawal GK, Rakwal R and Kim ST (2019) Proteomics of Rice—Magnaporthe oryzae Interaction: What Have We Learned So Far? Front. Plant Sci. 10:1383. doi: 10.3389/fpls.2019.01383
3. Halder, T., Choudhary, M., Liu, H., Chen, Y., Yan, G., & Siddique, K. H. M. (2022). Wheat Proteomics for Abiotic Stress Tolerance and Root System Architecture: Current Status and Future Prospects. Proteomes, 10(2), 17. https://doi.org/10.3390/proteomes10020017
4. John Momo, Abdul Rawoof, Ajay Kumar, Khushbu Islam, Ilyas Ahmad and Nirala Ramchiary (2022) Proteomics of Reproductive Development, Fruit Ripening, and Stress Responses in Tomato. https://doi.org/10.1021/acs.jafc.2c06564
Read More
The Fine Art of Protein Methylation: Mechanisms, Impacts, and Analytical Techniques
Maximizing Proteomic Potential: Top Software Solutions for MS-Based Proteomics


