How Age-Related Lipid Metabolism Drives Ovarian Cancer: A Multi-Omics Breakthrough
We are excited to highlight a groundbreaking study titled “Regulation of Age-Related Lipid Metabolism in Ovarian Cancer” published in the International Journal of Molecular Sciences (2025). This research employed RNA sequencing and quantitative lipidomics to investigate how aging reshapes the tumor microenvironment (TME) in ovarian cancer (OC). By analyzing gonadal adipose tissues from young and aged rat xenograft models, the study uncovered significant age-driven changes in lipid metabolism, immune cell dynamics, and gene expression that fuel tumor growth. The work underscores the critical role of lipidomics in decoding metabolic vulnerabilities in aging-associated cancers, offering novel therapeutic avenues for older OC patients.
Unraveling the Aging-Ovarian Cancer Nexus: Why Lipid Metabolism Matters
Ovarian cancer remains the deadliest gynecological malignancy, with advanced age being the strongest risk factor. Older patients face poorer prognoses due to unclear mechanisms linking aging to cancer progression. Emerging evidence suggests that the tumor microenvironment, particularly adipose tissue, plays a pivotal role in OC metastasis. However, how age-related metabolic shifts in adipose tissue drive tumor aggressiveness has been poorly understood.
This study bridges this gap by integrating transcriptomics and lipidomics to dissect the molecular and metabolic landscape of aged adipose TME. By comparing young and aged rat models, the authors identified key lipid species and inflammatory pathways that promote OC proliferation. Such insights are vital for developing targeted therapies tailored to older populations, where conventional treatments often fail.
Aged Adipose Tissue Accelerates Tumor Growth
Using a rat xenograft model, researchers observed that 100% of aged rats developed ovarian tumors, compared to 68.75% of young rats, with tumors in aged hosts being significantly larger (p = 0.002). RNA-seq revealed 2,323 differentially expressed genes (DEGs) in aged pre-tumor adipose tissue, including pro-inflammatory markers S100a8 and S100a9. Lipidomics further showed elevated free fatty acids (FFAs) and triglycerides (TGs) in aged rats, suggesting a lipid-rich TME conducive to cancer progression.
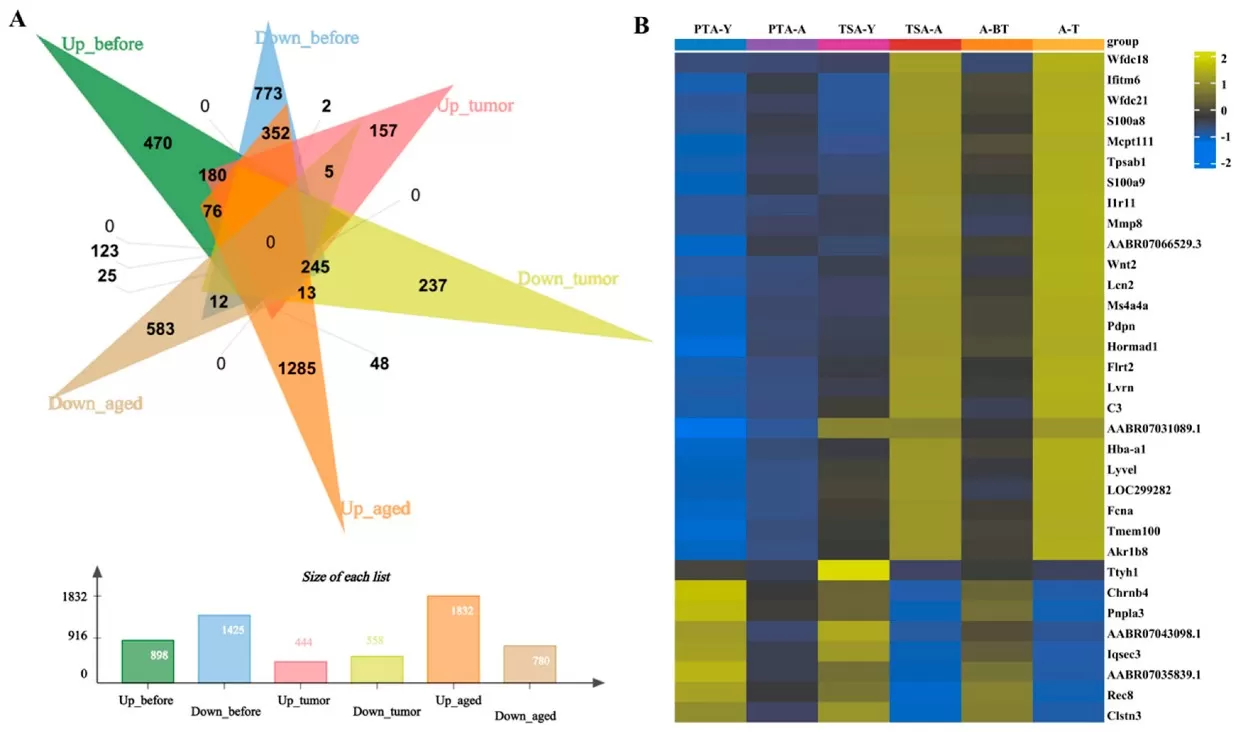
Transcriptome analysis shows differential gene expression in PTA-A&Y, TSA-A&Y, and PTA&TSA-A.
Lipidomic Shifts and Immune Dysregulation
Quantitative lipidomics identified 37 differentially expressed metabolites in aged adipose tissues, with omega-5 FFA (18:3) levels notably reduced. This lipid exhibited tumor-suppressive effects in vitro, inhibiting OC cell proliferation (p < 0.001). Concurrently, aged TMEs showed heightened infiltration of neutrophils, CD4+ T cells, and activated mast cells, linked to S100a8/S100a9 upregulation. These findings highlight a crosstalk between lipid metabolism and immune dysfunction in aging.
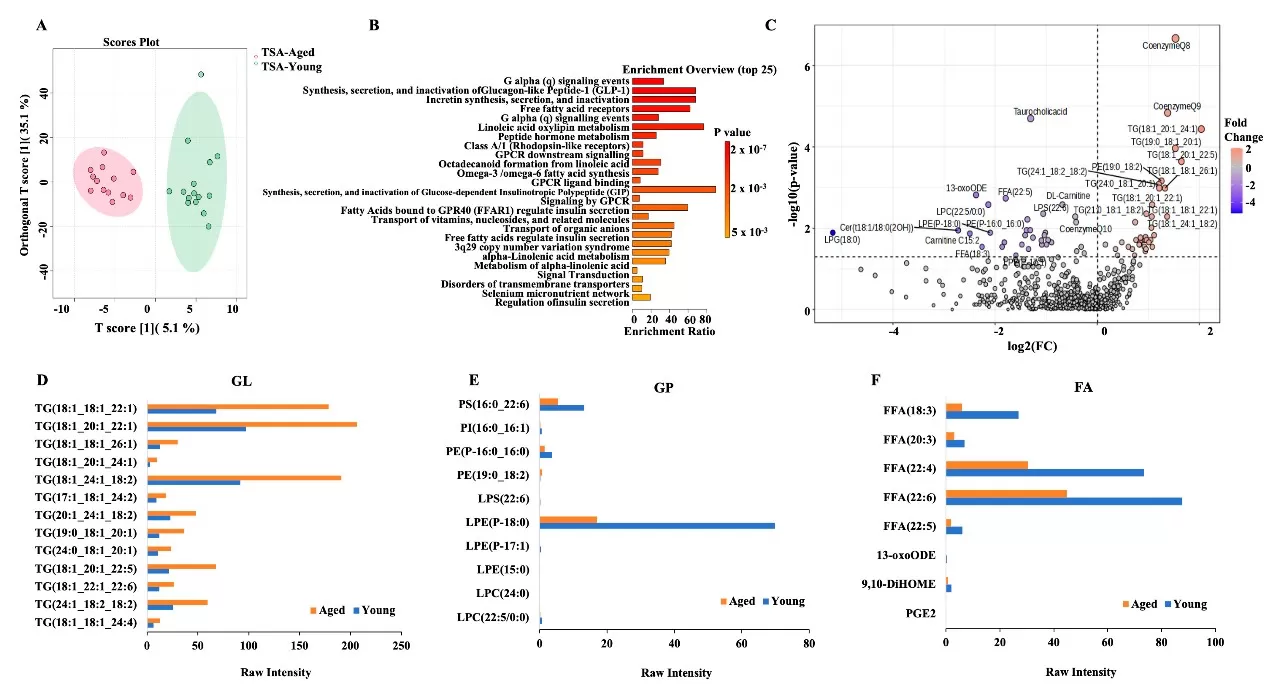
Differential expression of metabolites in TSA-A&Y adipose.
Gene-Lipid Networks in Cancer Promotion
Integrative analysis revealed strong correlations between S100a8/S100a9 expression, neutrophil infiltration, and FFA/TG levels. For instance, FFA (18:3) inversely correlated with tumor aggressiveness, while Pnpla3 (a lipid metabolism gene) was downregulated in aged rats. These networks position lipid metabolism as a central regulator of age-driven OC, offering biomarkers for therapeutic targeting.
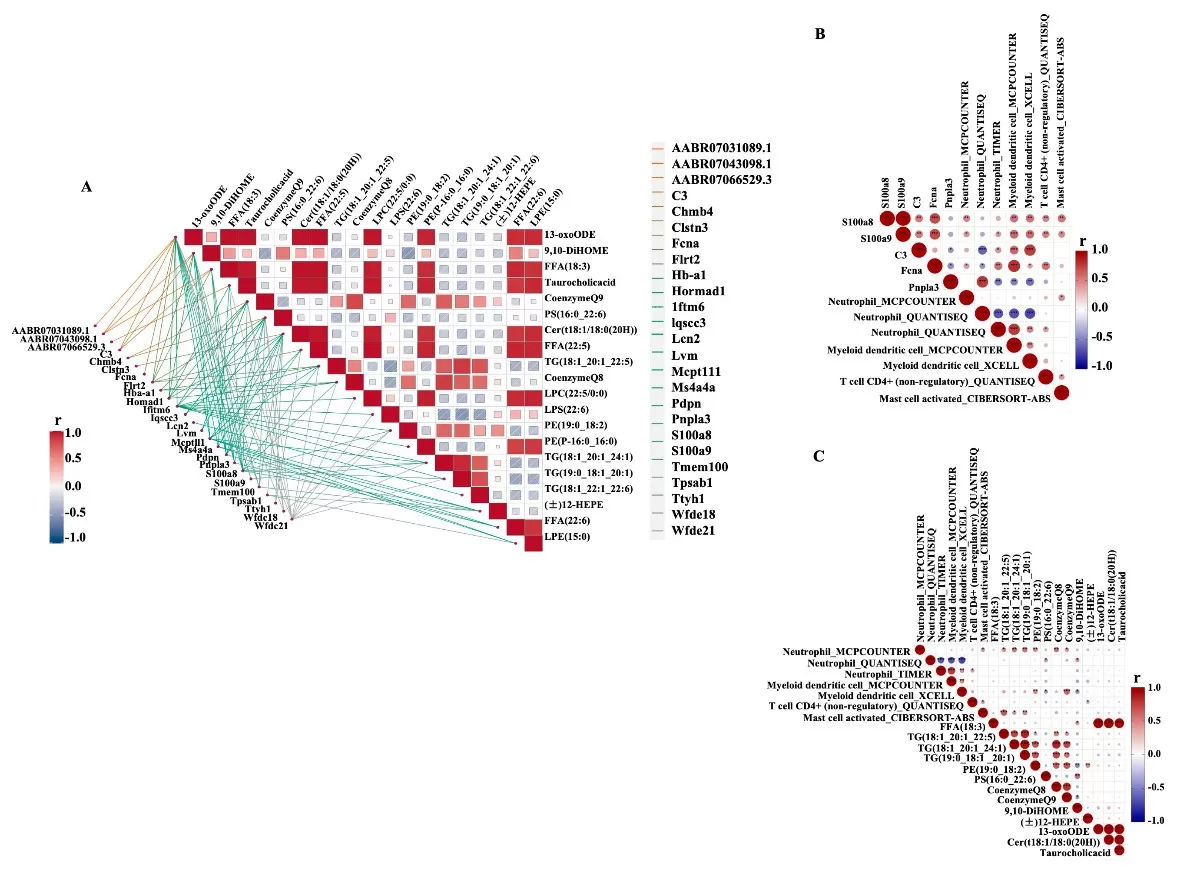
Transcriptome–metabolome correlative regulation.
Decoding Aging’s Role in Cancer: The Power of Multi-Omics
This study pioneers a holistic view of how aging reprograms the ovarian cancer microenvironment through lipidomic and transcriptomic synergy. The discovery of omega-5 FFA (18:3) as a tumor suppressor and S100a8/S100a9 as inflammatory drivers underscores the potential of lipid-focused therapies. Notably, the integration of lipidomics with immune profiling provides a blueprint for understanding metabolic-immune crosstalk in other aging-related cancers.
Metabolomics and lipidomics are indispensable for uncovering such mechanisms. These technologies enable precise mapping of metabolic pathways altered by aging, offering actionable targets for intervention. Future studies could explore dietary or pharmacological modulation of FFAs or leverage single-cell lipidomics to dissect adipocyte heterogeneity in the TME.
Unlock Precision Insights with Metware’s Lipidomics Solutions
At Metware Biotechnology, we specialize in quantitative lipidomics and multi-omics integration to empower groundbreaking cancer research. Our platform detects over 4,000 lipid species with high sensitivity, enabling researchers to uncover metabolic drivers of disease. Coupled with RNA-seq, our services provide end-to-end solutions for decoding complex biological systems.
Why Choose Metware?
- Cutting-edge LC-MS/MS technology for unparalleled accuracy.
- Expert bioinformatics to unravel gene-metabolite networks.
- Tailored solutions for preclinical and clinical studies.


