Unveiling AFADESI Spatial Metabolomics
In the realm of mass spectrometry imaging (MSI), matrix-assisted laser desorption/ionization (MALDI) and desorption electrospray ionization (DESI) are two cornerstone techniques. While MALDI relies on matrix coating and risks sample damage, DESI’s matrix-free design and non-contact ionization mode simplify sample preparation and preserve tissue integrity, making it a preferred choice for spatially resolved metabolomics in biomedical research.
Air Flow-Assisted Desorption Electrospray Ionization (AFADESI) is an innovative MSI technology that integrates laminar airflow with a quartz transfer tube to establish an ion transmission channel. This setup enables charged droplets to undergo desolvation and ion focusing within a 5–10 cm transmission distance, boosting ionization efficiency by 3–5 times. By dynamically adjusting airflow rates (0.5–2 L/min) and spray voltages (3–5 kV), AFADESI adapts to tissue hardness and metabolite polarity variations. Coupled with Orbitrap-based high-resolution mass spectrometry (HRMS) and deep learning algorithms (e.g., ResNet for spatial feature extraction), AFADESI distinguishes spatial distribution patterns of over 2,000 metabolites, offering molecular topological insights for studies on tumor microenvironments and other heterogeneous systems.
High Coverage: Capturing Metabolic Diversity in Complex Tissues
AFADESI-MSI is a label-free, highly sensitive method for imaging polar small-molecule metabolites. It enables in situ spatial resolution of endogenous metabolites in complex tissues like the brain without extensive preprocessing. For instance, in rat brain studies, AFADESI simultaneously detected approximately 1,500 metabolites spanning three functional modules and 11 molecular classes:
1. Nitrogen Metabolism: Choline derivatives, polyamines, amino acids.
2. Energy Metabolism: Carnitines, nucleotides, organic acids, carbohydrates.
3. Lipid and Steroid Regulation: Cholesterol derivatives, bile acids, lipid species.
By constructing metabolite-spatial coordinate correlation maps, AFADESI revealed region-specific metabolic networks, such as glutamine enrichment in the hippocampus and polyamine gradients in the cortex, elucidating molecular mechanisms underlying neural functions.
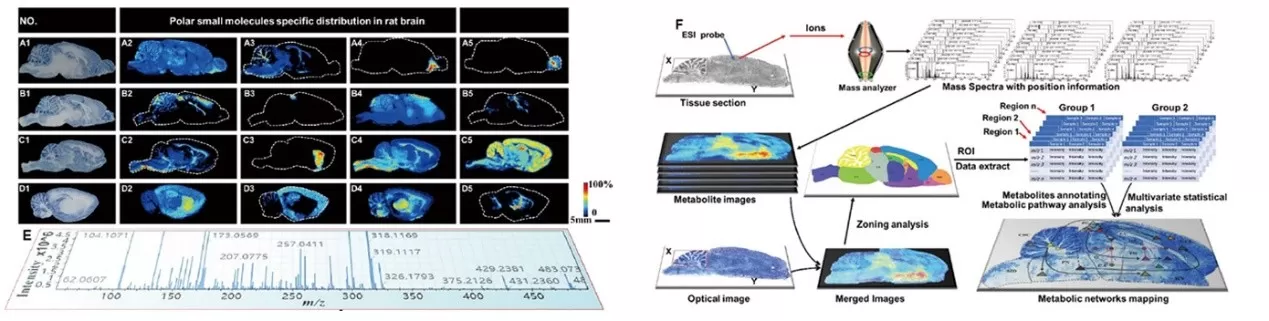
AFADESI-MSI workflow and representative ion images of polar endogenous metabolites in rat brain
Broad Dynamic Range: Visualizing Subtle Variations with Precision
AFADESI’s wide dynamic range allows precise detection of metabolites across concentration gradients. In a study of scopolamine-induced cognitive impairment in rats, histamine levels in the sagittal brain section showed a dramatic drop from 4.0×10⁴ (control) to 1.0×10³ (drug-treated group). Despite low abundance, AFADESI clearly visualized histamine’s distinct spatial distribution. This capability enhances image contrast and stratification, enabling accurate metabolite mapping in complex tissues.
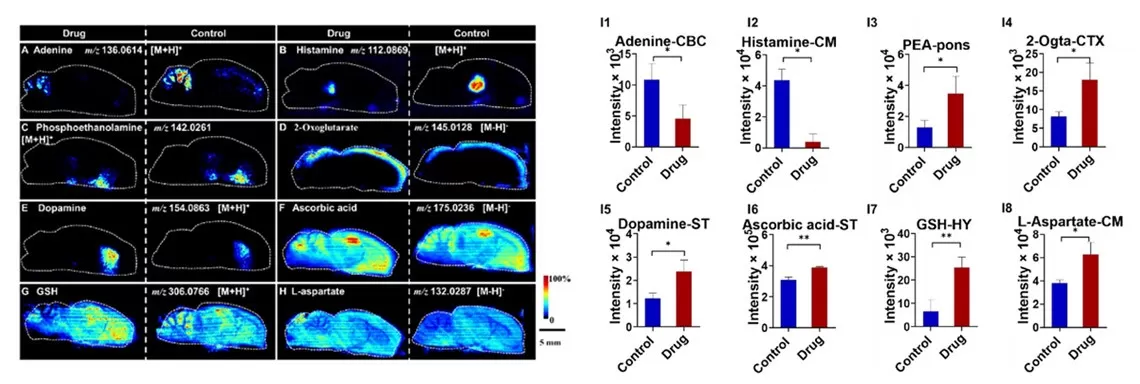
AFADESI-MSI images of rat brain under scopolamine treatment
Enhanced Metabolite Coverage via Polarity-Tuned Solvents
AFADESI’s flexibility extends to solvent selection. By tailoring spray solvents to match tissue morphology and metabolite polarity, researchers detected 2,174 metabolites and 311 lipids in rat knee cartilage, and 2,071 metabolites and 382 lipids in bone marrow tissues. This adaptability ensures comprehensive metabolite profiling across diverse biological matrices.
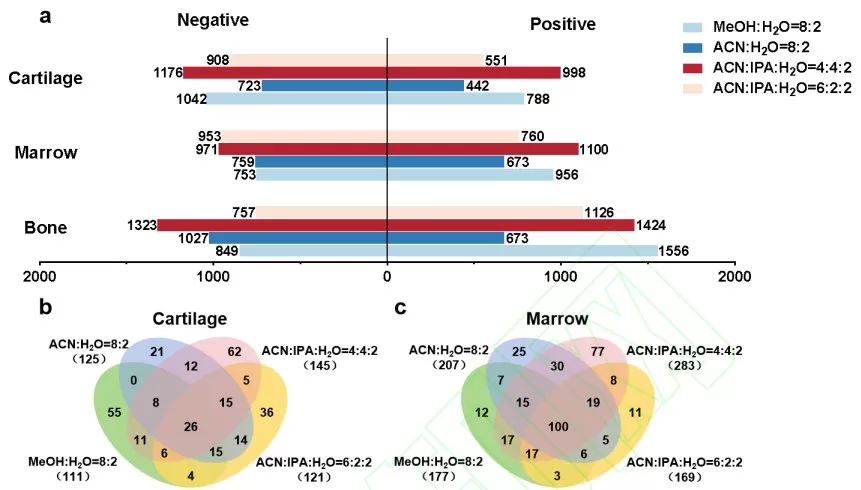
Metabolic and lipid coverage in rat knee cartilage and bone marrow
High Reproducibility: Consistent Spatial Mapping
AFADESI’s reliability was validated using six adjacent rat knee tissue slices. Under both positive and negative ion modes, low-, medium-, and high-intensity metabolites exhibited consistent spatial distributions and signal intensities across slices. The relative standard deviation (RSD) of ion intensities between adjacent slices remained below 20%, underscoring the method’s robustness.
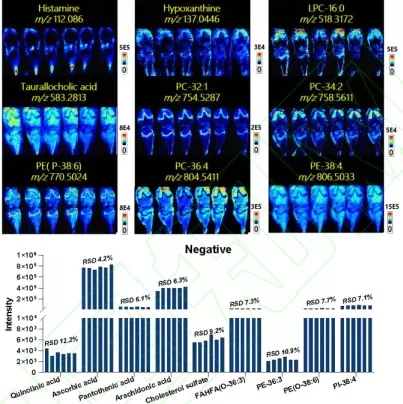
Reproducibility evaluation of AFADESI-MSI in sequential tissue sections.
High Throughput Metabolic Profiling: From Single Organs to Whole-Body Analysis
AFADESI covers metabolites across a broad mass range (m/z 50–2000), from polar small molecules (e.g., alanine, m/z 89) to complex lipids (e.g., phosphatidylinositol, m/z 1845). Leveraging this throughput, a multi-organ study identified 1,620 metabolites in the brain, liver, kidney, heart, spleen, lung, muscle, pancreas, and serum. Integration with LC-MS and adduct pattern analysis expanded the metabolite database to 27,407 ion adducts, enabling the first whole-body spatial metabolomic atlas in mice. This leap from hundreds to thousands of metabolites per organ marks a paradigm shift in systemic metabolic research.
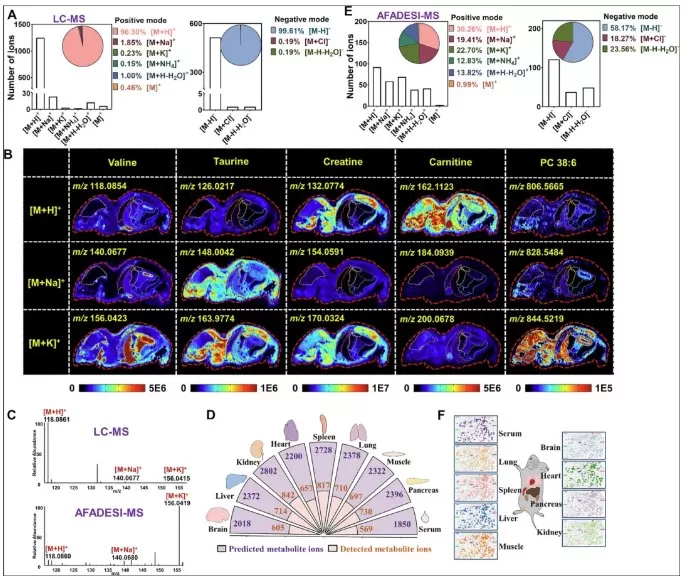
Whole-body metabolic mapping of mouse organs using AFADESI
Applications of AFADESI: Bridging Research and Clinical Translation
AFADESI’s strengths—in situ analysis, matrix-free operation, high sensitivity, and broad dynamic range—have fueled advancements in diverse fields:
- Disease Mechanisms: Unraveling tumor metabolic heterogeneity and neurotransmitter anomalies in neurodegenerative diseases.
- Drug Development: Tracking nanodrug release and mapping hepatorenal toxicity metabolites.
- Plant Science: Visualizing drought-induced stress molecules.
- Microbial Interactions: Imaging quorum-sensing signals in biofilms.
- Clinical Diagnostics: Intraoperative metabolic boundary delineation and biomarker discovery.
- Environmental Toxicology: Assessing organ-specific pollutant accumulation.
AFADESI spatial metabolomics, with its "in situ, label-free, high-throughput" capabilities, offers unparalleled molecular insights into biological systems. As the technology evolves, it will undoubtedly become a cornerstone of next-generation biomedical research.
References
Pang X, Gao S, Ga M, Zhang J, Luo Z, Chen Y, Zhang R, He J, Abliz Z. Mapping Metabolic Networks in the Brain by Ambient Mass Spectrometry Imaging and Metabolomics. Anal Chem. 2021 May 4;93(17):6746-6754. doi: 10.1021/acs.analchem.1c00467.
Zhu Y, Zang Q, Luo Z, He J, Zhang R, Abliz Z. An Organ-Specific Metabolite Annotation Approach for Ambient Mass Spectrometry Imaging Reveals Spatial Metabolic Alterations of a Whole Mouse Body. Anal Chem. 2022 May 24;94(20):7286-7294. doi: 10.1021/acs.analchem.2c00557.
Read more
- Spatial Metabolomics: Transforming Biomedical and Agricultural Research
- Spatial Metabolomics Explained: How It Works and Its Role in Cancer Research
- MALDI, DESI, or SIMS? How to Choose the Best MSI Techniques for Spatial Metabolomics
- How to Prepare Samples for Spatial Metabolomics: The Essential Guide You Need
- Unlocking Precision in Spatial Metabolomics: Essential Detection Parameters for Cutting-Edge Research


