Unlocking Protein Purity: Advanced Dialysis Techniques for Precision in Protein Research
Dialysis is a cornerstone technique in protein purification, enabling the removal of small-molecule impurities such as salts, detergents, and metabolites while retaining target proteins. This blog explores the principles, optimized workflows, and specialized strategies of dialysis, offering actionable insights for researchers to enhance efficiency and reproducibility in protein studies.
1. Principles of Dialysis
Dialysis leverages the selective permeability of semipermeable membranes to separate molecules based on size. The core mechanisms include:
Diffusion-Driven Exchange: Small molecules passively diffuse from high-concentration regions (sample) to low-concentration regions (dialysate).
Molecular Weight Cutoff (MWCO): Membranes with defined pore sizes (e.g., 3.5–100 kDa) retain proteins above the MWCO while allowing smaller molecules to pass.
Dynamic Equilibrium: The process concludes when the concentrations of small molecules equilibrate across the membrane.
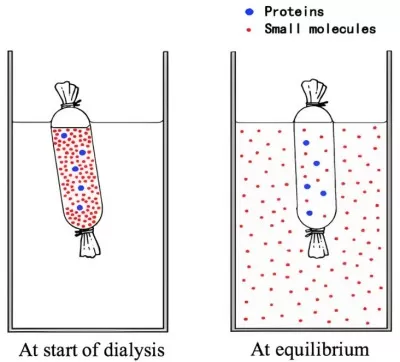
Protein Dialysis
2. Workflow and Technical Details
2.1 Membrane Selection and Pretreatment
Material Properties:
- Regenerated Cellulose (RC): High mechanical strength, uniform pores, but requires pre-wetting to minimize protein adsorption (e.g., Spectra/Por® 6, MWCO 1–50 kDa).
- Cellulose Acetate (CA): Hydrophilic and ideal for viscous samples (e.g., glycerol-containing buffers), but less resistant to harsh chemicals.
- Low-Protein-Binding Membranes: Chemically modified (e.g., PEG-coated CE membranes) to reduce nonspecific adsorption (<5 μg/cm²).
MWCO Optimization:
- Empirical rule: Select a membrane with an MWCO 1/3–1/5 of the target protein’s molecular weight (e.g., 6–10 kDa MWCO for a 30 kDa protein).
- Fibrous vs. Globular Proteins: Fibrous proteins (e.g., collagen) may require lower MWCO membranes due to their extended conformations.
Pretreatment Protocols:
- Deionized Water Soaking: 30 minutes to remove preservatives (e.g., Na₂S).
- EDTA Treatment: Chelates metal ions to prevent pore clogging (1 mM EDTA, pH 8.0).
- Ethanol Activation: Enhances hydrophilicity of hydrophobic membranes (50% ethanol, followed by dialysate rinse).
2.2 Dialysis Setup and Leak Prevention
Closure Methods:
1) Clamps: Polypropylene clamps (e.g., Spectra/Por® Closure) ensure uniform sealing.
2) Heat Sealing: For heat-sealable membranes (e.g., Float-A-Lyzer® G2) at 80°C for 2 seconds.
Volume Control: Fill dialysis bags to ≤80% capacity to accommodate expansion, especially for high-salt or glycerol-containing samples.
2.3 Dialysis Buffer Design
Ionic Strength Gradients: Gradually reduce salt concentrations (e.g., 1 M → 50 mM NaCl) in ≤50% increments to prevent protein precipitation.
pH Stability:
- Tris-HCl: Suitable for pH 7–9 (precool to offset temperature-dependent pH shifts).
- Phosphate Buffers: Avoid with Ca²⁺-containing proteins to prevent precipitation.
Additives:
Reducing Agents: DTT (1–5 mM, replenished every 4 hours) or TCEP (0.5–2 mM, stable for prolonged dialysis).
Detergents: Use nonionic detergents (e.g., 0.1% Triton X-100) below the membrane’s MWCO. For retained detergents (e.g., DDM), maintain concentrations above the critical micelle concentration (CMC) in the dialysate.
3. Advanced Techniques
Temperature and Stirring Optimization:
- Cold Dialysis: Use a recirculating chiller (±0.5°C) to maintain 4°C.
- Magnetic Stirring: PTFE-coated stir bars at 200–300 rpm enhance mixing without damaging membranes.
Pressure-Assisted Dialysis:
- Positive Pressure: Stirred cell systems
- Negative Pressure: Vacuum-assisted systems
4. Specialized Applications
4.1 Membrane Protein Dialysis
Detergent Management:
1) Balance detergent concentrations (e.g., 0.03% DDM) above CMC to prevent aggregation.
2) Gradual detergent exchange (e.g., 1% OG → 0.05%) preserves solubility.
Liposome Co-Dialysis: Incorporate lipid bilayers (e.g., DOPC/cholesterol) to mimic native membrane environments.
4.2 Large Complexes and Viral Particles
- Ultra-Low MWCO Membranes: 100 kDa membranes for buffer exchange of viral capsids (e.g., adenovirus) or protein polymers.
- Tangential Flow Filtration (TFF): Combines dialysis with continuous concentration using 100 kDa cassettes.
4.3 Isotope-Labeled Proteins
- Isotope Retention: Use deuterated or stable isotope-enriched buffers (e.g., D₂O-based) to prevent dilution.
- Sealed Systems: Operate in glove boxes to minimize isotopic exchange with ambient air.
5. Troubleshooting Common Issues
5.1 Post-Dialysis Precipitation
Causes: Sudden pH or ionic strength changes.
Solutions:
- Gradient Dialysis: Incrementally adjust parameters (≤50% salt reduction or ≤0.5 pH unit shifts per step).
- Additives: Arginine (0.5–1 M) or sucrose (5–10%) enhance solubility.
5.2 Protein Loss via Membrane Adsorption
Quantification: BCA assay to measure preand post-dialysis concentrations (RC: 3–10% loss; CE: <2%).
Mitigation:
- Preblocking: Treat membranes with 1% BSA or 0.1% Tween-20 for 1 hour.
- Competitive Agents: Add 0.01% CHAPS or 0.1 mg/mL heparin to samples.
6. Comparative Analysis of Desalting Techniques
Dialysis excels in preserving protein native states but requires careful optimization for time-sensitive projects. Ultrafiltration is preferable for rapid desalting, while desalting columns suit high-throughput workflows.
Comparison of four desalting techniques
|
Technique |
Advantages |
Limitations |
Ideal Use Cases |
|
Dialysis |
Gentle, cost-effective, scalable |
Time-consuming, frequent buffer changes |
Buffer exchange, large-volume samples |
|
Ultrafiltration |
Rapid, simultaneous concentration |
Shear stress may denature proteins |
Fast desalting, small-to-medium volumes |
|
Desalting Columns |
Extremely fast (5–10 minutes) |
Limited capacity (<5 high="" p=""> |
Small-volume, high-throughput needs |
|
Dilution-UF |
Prevents precipitation |
Multi-step complexity |
High-salt samples requiring dilution |
<5 high="" p="">
<5 high="" p="">
<5 high="" p="">
Dialysis remains indispensable in protein purification, balancing simplicity with versatility. By mastering membrane selection, buffer design, and advanced techniques, researchers can tailor dialysis workflows to diverse applications—from membrane protein studies to isotope labeling. As automation and material science advance, dialysis will continue evolving, bridging classical methods with next-generation proteomics.


