A Guide to Protein Database Selection
Proteomics relies on protein databases for spectrum prediction and comparison with mass spectrometry data to achieve protein identification. Therefore, protein databases serve as the foundation of proteomic analysis, and their completeness and accuracy directly impact the quality of final proteomic data.
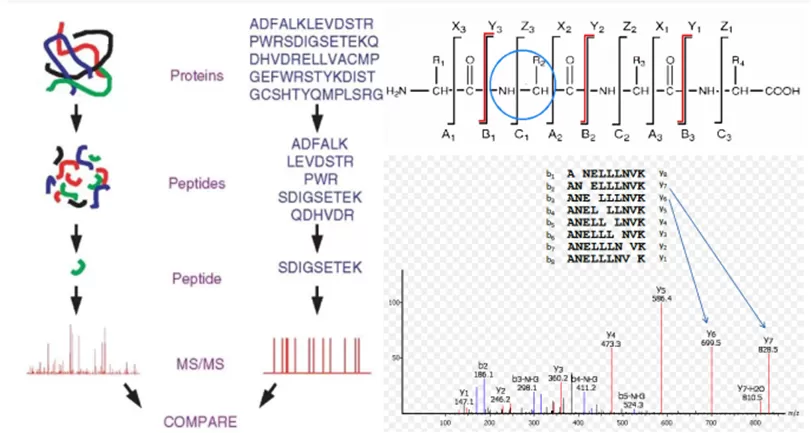
Figure 1. The princeple of protein identification
Human Proteome
Compared to other species, research on the human proteome is relatively well-established, with the commonly utilized database provided by UniProt. Within UniProt, there are three sub-databases dedicated to the human proteome: UniProtKB/Swiss-Prot (referred to as Swiss-Prot), Proteome (UP000005640), and UniProtKB (comprising Swiss-Prot and TrEMBL). These databases differ in terms of protein count, accuracy, and annotation depth.
|
UniProt |
Type |
Total Number of Protein Sequences |
Number of Unique Protein Sequences |
|
UniProtKB |
Swiss-Prot(Reviewed) |
20404 |
20330 |
|
TrEMBL(Unreviewed) |
186900 |
|
|
|
Total |
207304 |
182025 |
|
|
Proteome |
Swiss-Prot(Reviewed) |
20389 |
一 |
|
TrEMBL(Unreviewed) |
61448 |
一 |
|
|
Total |
81837 |
81579 |
Swiss-Prot stands out among these databases. It is a high-quality, manually curated, non-redundant database primarily derived from research findings in literature and computationally analyzed results validated by E-value verification. Only data meeting quality standards are included in this database, making it a verified resource.
UniProtKB/TrEMBL, in contrast, consists of automatically translated nucleotide-encoded sequences, which undergo high-quality annotation and classification. This database is categorized as unverified.
Proteomes contain protein information translated and annotated from nucleotide sequences of whole-genome sequenced species, with each dataset assigned a Unique Proteome Identifier (UPID).
1) Comparison of Detected Protein Count
To assess the influence of different databases on the quality of proteomic data, Metware conducted searches using human cell proteomic data (divided into groups A and B, each with 4 replicates) across various databases. Subsequently, the qualitative and quantitative results of protein identification were evaluated.
Peptides identified using the Swiss-Prot, Proteome, and UniProteinKB databases numbered 100,821, 100,723, and 99,597, respectively. The corresponding quantities of identified proteins were 7,798, 8,158, and 8,452, respectively. The proportion of proteins and peptides identified across all three databases collectively was 85.97% and 87.21%, respectively.
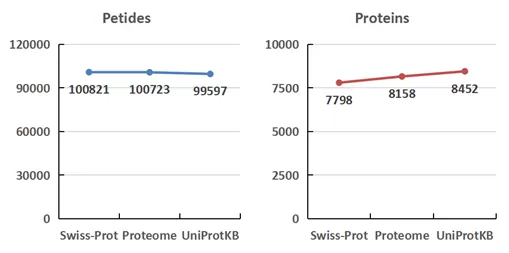
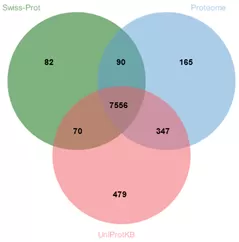
Figure 2. Differences in Protein Detection Data Across Different Databases for Cell Samples
2) Comparison of Missing Values
An analysis was conducted on the quantitative missing values across different databases. The trend of missing values for all samples showed consistency across the three databases. However, Proteome and UniProtKB exhibited overall higher missing value rates compared to Swiss-Prot. In the comparison of missing values, Swiss-Prot showed a slight advantage over the other databases.
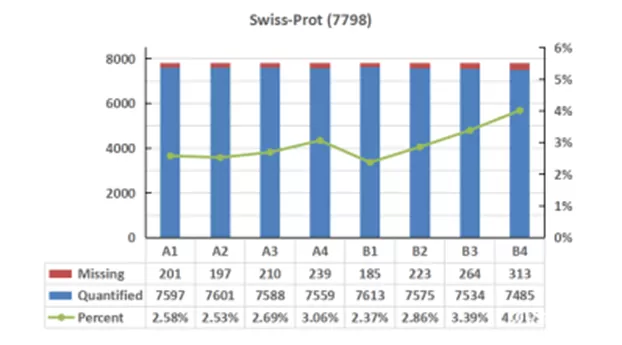
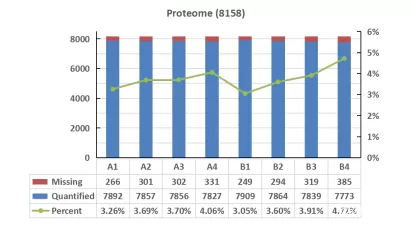
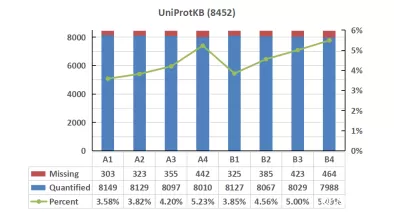
Figure 3. Missing Values in Protein Detection across Different Databases for Cell Samples
Summary:
a) Compared to the UniProtKB/Swiss-Prot database, using the Proteome and UniProtKB databases for searching resulted in a slight increase in the number of identified proteins by 4.62% and 8.44%, respectively. However, the increase was not significant.
b) Compared to the Proteome and UniProtKB databases, the Swiss-Prot database had the least sequence information and identified the fewest proteins. However, it had the highest number of identified peptide segments, indicating higher accuracy and better matching of protein sequences in Swiss-Prot.
c) With the increase in database usage, the proportion of missing values in quantified proteins also increased, indicating that many of the additionally identified proteins may have lower ion intensity and could potentially be false positives relative to the database.
In summary, while the Swiss-Prot database may have slightly fewer identified proteins, it offers superior qualitative accuracy and quantitative stability. Overall, for human cell samples, it is recommended to use the Swiss-Prot database for proteomic analysis.


