Transcriptomics + Proteomics + Metabolomics
Transcriptomics + Proteomics + Metabolomics
Technology Introduction to Transcriptomics+Proteomics + Metabolomics
The transcriptome provides crucial insights into gene expression within a biological system, while the proteome offers a comprehensive overview of the expressed proteins, including their modifications and interactions. The metabolome serves as the direct readout of the system’s phenotype, with metabolites representing the final products of gene transcription and expression, influenced by internal and external regulation, forming the foundational basis of the observed phenotype. Together, these three omics layers—transcriptomics, proteomics, and metabolomics—offer a holistic view of biological processes, linking gene expression to protein activity and metabolic outcomes.
Correlating transcriptomic data, which often includes a large number of differentially expressed genes, with differential proteins or protein modifications detected by proteomics, as well as differential metabolites identified by metabolomics, can pinpoint key genes, proteins, protein modifications, metabolites, and metabolic pathways closely associated with internal system changes. This integrated approach facilitates a more comprehensive understanding of biological problems.
MetwareBio offers integrated transcriptomics, proteomics, and metabolomics analysis services, enabling the normalization, comparative analysis, and correlation of bulk data from these omics layers to establish relationships between different molecular layers. Coupled with GO functional analysis, metabolic pathway enrichment, and molecular interaction analyses, it provides a thorough understanding of biomolecular functions and regulatory mechanisms. Integration of transcriptomics, proteomics, and metabolomics allows for mutual validation and complementary insights, resulting in a holistic understanding of biological changes.
Appications of Transcriptomics + Proteomics + Metabolomics
Project Workflow of Proteomics + Metabolomics
Proteomics + Metabolomics Case Study
Abstract:
Hepatic ischemia–reperfusion (IR) injury is the primary reason for complications following hepatectomy and liver transplantation (LT). Insulin-induced gene 2 (Insig2) is one of several proteins that anchor the reticulum in the cytoplasm and is essential for metabolism and inflammatory responses. However, its function in IR injury remains ambiguous. In this study, Insig2 global knock-out (KO) mice and mice with adeno-associated-virus8 (AAV8)-delivered Insig2 hepatocyte-specific overexpression were subjected to a 70% hepatic IR model. Liver injury was assessed by monitoring hepatic histology, inflammatory responses, and apoptosis. Hypoxia/reoxygenation stimulation (H/R) of primary hepatocytes and hypoxia model induced by cobalt chloride (CoCl2) were used for in vitro experiments. Multi-omics analysis of transcriptomics, proteomics, and metabolomics was used to investigate the molecular mechanisms underlying Insig2. Results revealed hepatic Insig2 expression was significantly reduced in clinical samples undergoing LT and the mouse IR model. Our findings showed that Insig2 depletion significantly aggravated IR-induced hepatic inflammation, cell death and injury, whereas Insig2 overexpression caused the opposite phenotypes. The results of in vitro H/R experiments were consistent with those in vivo. Mechanistically, multi-omics analysis revealed that Insig2 is associated with increased antioxidant pentose phosphate pathway (PPP) activity. The inhibition of glucose-6-phosphate-dehydrogenase (G6PD), a rate-limiting enzyme of PPP, rescued the protective effect of Insig2 overexpression, exacerbating liver injury. Finally, our findings indicated that mouse IR injury could be attenuated by developing a nanoparticle delivery system that enables liver-targeted delivery of substrate of PPP (glucose 6-phosphate). In conclusion, Insig2 has a protective function in liver IR by upregulating the PPP activity and remodeling glucose metabolism. The supplementary glucose 6-phosphate (G6P) salt may serve as a viable therapeutic target for alleviating hepatic IR.
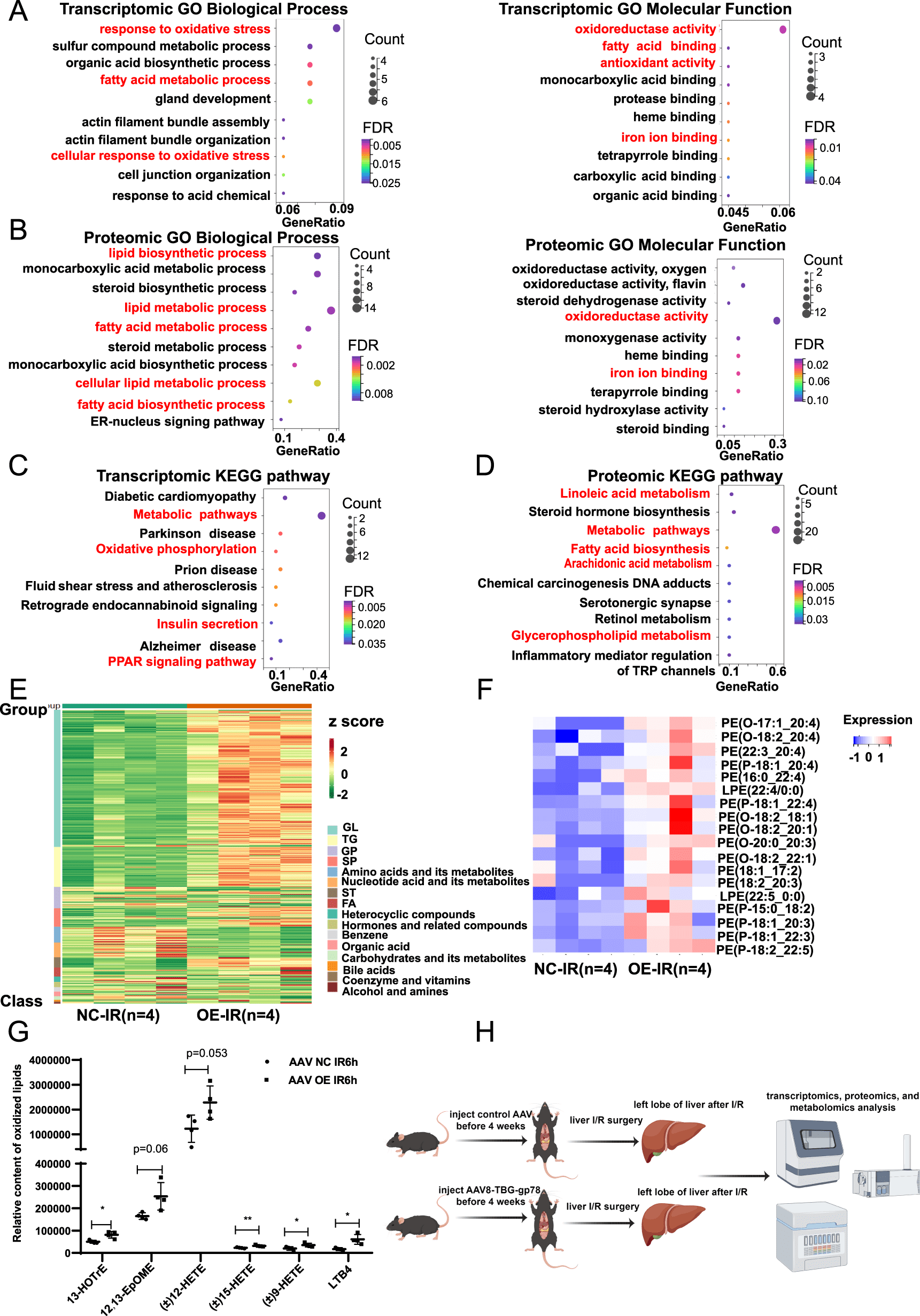
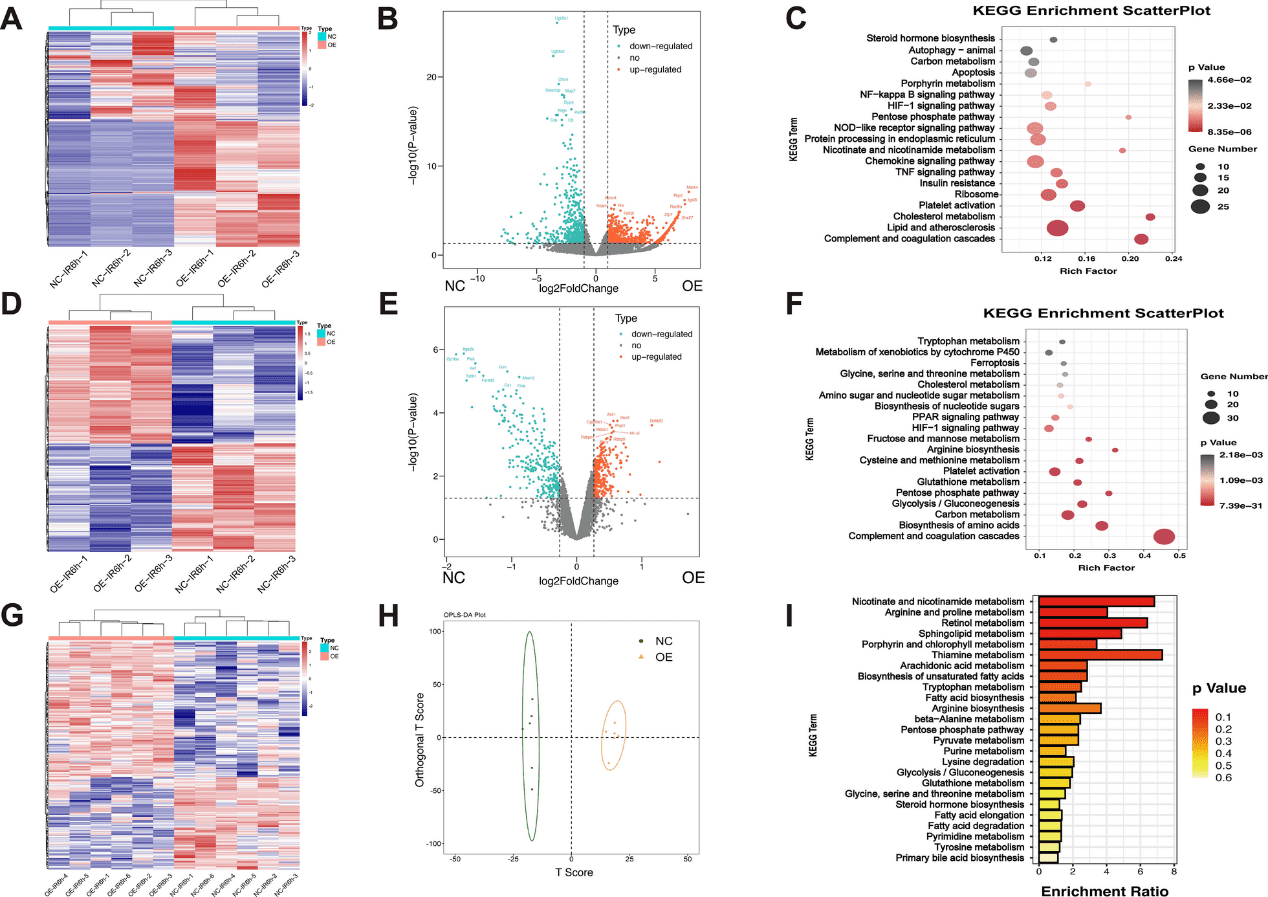 Transcriptome, proteome, and metabolome network analysis reveals disturbed pathways in Insig2 overexpression mice relative to those in WT controls subjected to IR injury
Transcriptome, proteome, and metabolome network analysis reveals disturbed pathways in Insig2 overexpression mice relative to those in WT controls subjected to IR injury
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.