Proteomics + Metabolomics
Proteomics + Metabolomics
Technology Introduction to Proteomics + Metabolomics
With the advancement of high-throughput sequencing technologies, the application of ultra-high-resolution mass spectrometers, the development of new statistical tools and computational methods, and the continuous enrichment of omics databases, omics technologies are evolving from traditional single-omics to multi-omics approaches. Multi-omics-driven systems biology research is poised to lead the next wave of discoveries in life sciences.
MetwareBio offers integrated proteomics and metabolomics analysis services, enabling the normalization, comparative analysis, and correlation analysis of bulk data from both proteomic and metabolomic layers. This comprehensive approach establishes data relationships between different molecular layers. Coupled with GO functional analysis, metabolic pathway enrichment, and molecular interaction analyses, it provides a thorough understanding of biomolecular functions and regulatory mechanisms. By integrating proteomics and metabolomics, we achieve mutual validation and complementary insights, resulting in a holistic understanding of biological changes. This approach proposes models for molecular biological change mechanisms, identifies key metabolic pathways, proteins, and metabolites for further in-depth experimental analysis and application, and brings new highlights and perspectives to research.
Appications of Proteomics + Metabolomics
Project Workflow of Proteomics + Metabolomics
Proteomics + Metabolomics Case Study
Abstract:
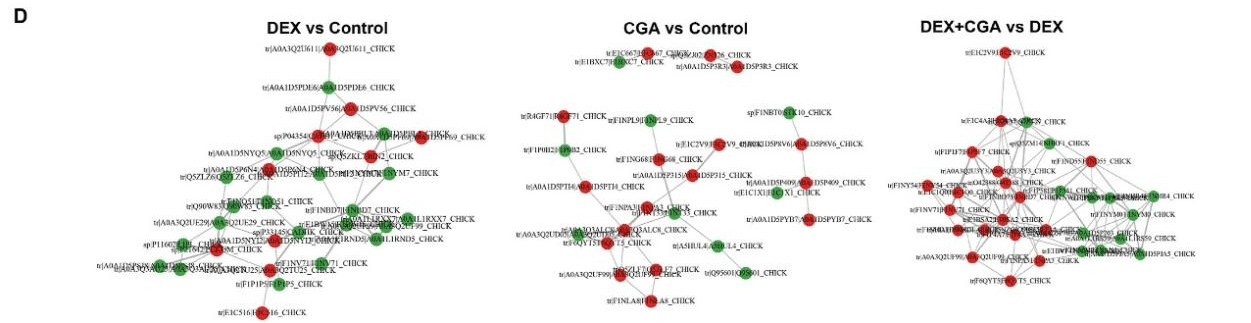
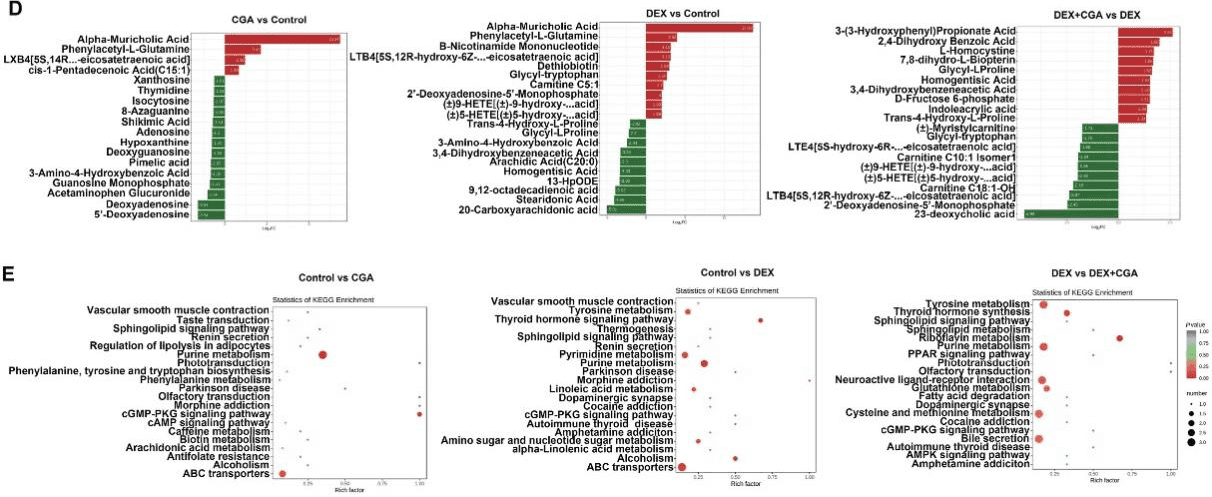
Intensive production can cause immunological stress in commercial broilers. Chlorogenic acid (CGA) regulates the intestinal microbiota, barrier function, and immune function in chickens. As complex interrelations regulate the dynamic interplay between gut microbiota, the host, and diverse health outcomes, the aim of this study was to elucidate the immunoregulatory mechanisms of CGA using multi-omics approaches. A total of 240 one-day-old male broilers were assigned to a 2 × 2 factorial design with 2 CGA levels (0 or 500 mg/kg) either with or without dexamethasone (DEX) injection for a 21-day experimental period. Therefore, there were 4 dietary treatments: control, DEX, CGA, and DEX + CGA, with 6 replicates per treatment. CGA supplementation improved (P < 0.05) growth performance, jejunal morphology, jejunal barrier function, and immune function in DEX-treated broilers. Moreover, in DEX + CGA-treated broilers, the increase in gut microbiome diversity (P < 0.05) was consistent with a change in taxonomic composition, especially in the Clostridiales vadin BB60_group. Additionally, the levels of short-chain fatty acids increased remarkably (P < 0.01) after CGA supplementation. This was consistent with the Kyoto Encyclopedia of Genes and Genomes analysis results that the “pyruvate fermentation to butanoate” pathway was more enriched (P < 0.01) in the DEX + CGA group than in the DEX group. Proteomics revealed that CGA treatment increased the expression of several health-promoting proteins, thymosin beta (TMSB4X) and legumain (LGMN), which were verified by multiple reaction monitoring. Metabolomics revealed that CGA treatment increased the expression of health-promoting metabolites (2,4-dihydroxy benzoic acid and homogentisic acid). Proteomic and metabolic analyses showed that CGA treatment regulated the peroxisome proliferator-activated receptor (PPAR) and mitogen-activated protein kinase (MAPK) pathways. Western blotting results support these findings. Pearson’s correlation analyses showed correlations (P < 0.01) between altered immune function, jejunal barrier function, different microbiota, proteins, and metabolites parameters. Overall, our data indicate that CGA treatment increased growth performance and improved the immunological functions of DEX-treated broilers by regulating gut microbiota and the PPAR and MAPK pathways. The results offer novel insights into a CGA-mediated improvement in immune function and intestinal health.
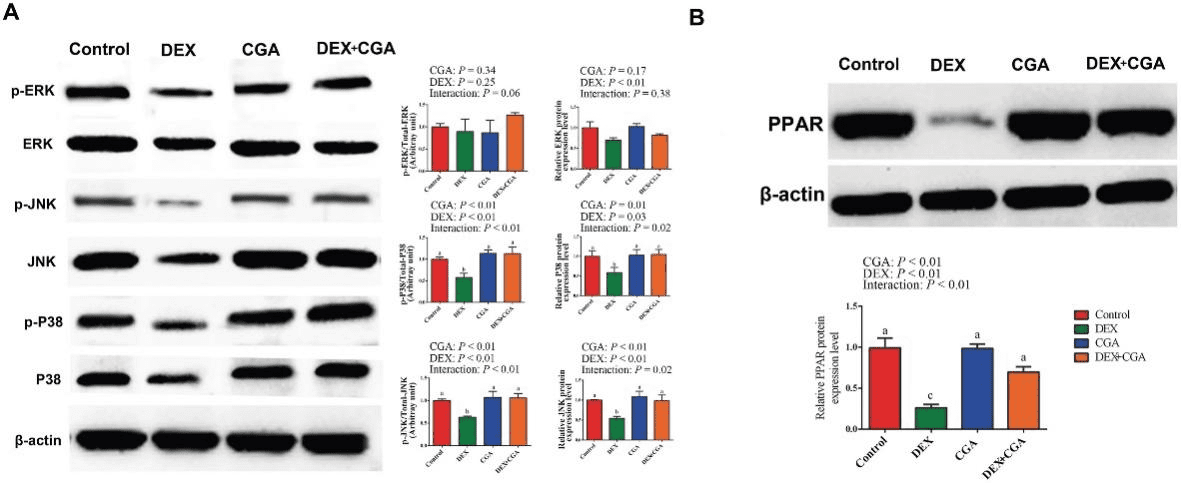 Analysis of proteomics, metabolomics and experimental validation
Analysis of proteomics, metabolomics and experimental validation
Abstract:
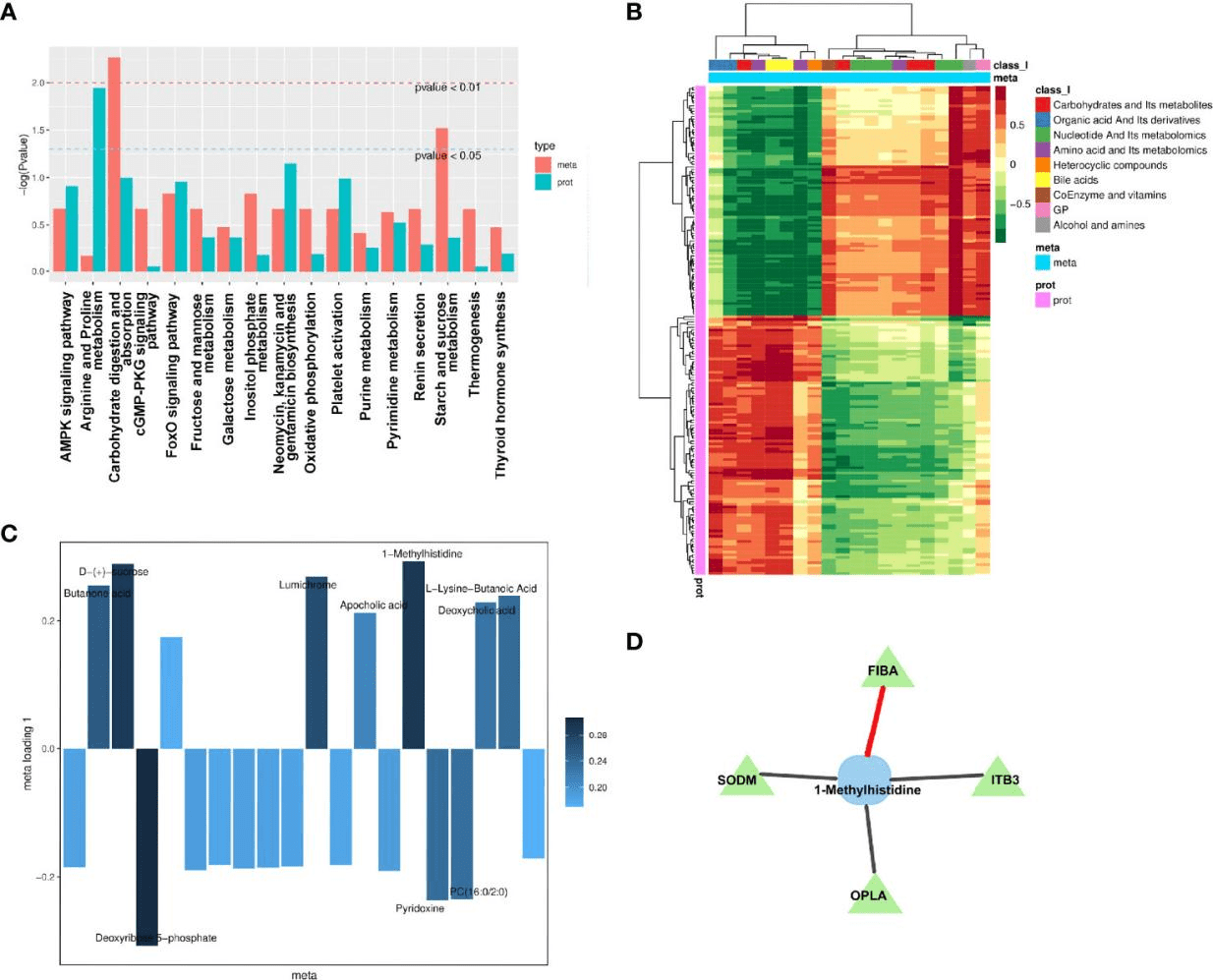
Cholelithiasis is a gastrointestinal disease that is associated with the highest socioeconomic cost. A diagnosis of cholelithiasis based on clinical features is significantly limited, and direct molecular insights into cholelithiasis and the relationship between cholelithiasis and clinical biochemical parameters are unclear. The aim of this study is to uncover direct molecular insights into cholelithiasis and the relationship between cholelithiasis and clinical biochemical parameters, then identify sensitive and specific biomarkers for this disease. In this study, parallel metabolomic and proteomic analyses of plasma from cholelithiasis patients (CPs) and healthy control individuals (HCs) without cholelithiasis were performed using ultrahigh-performance liquid chromatography-tandem mass spectrometry. A multimodule coexpression network analysis and integrated machine learning methods, including least absolute shrinkage and selection operator, random forest, and support vector machine, were used for bioinformatic analyses. An independent cohort and the cross-validation of the combination of two cohorts were used to evaluate the diagnostic performance of the panel. The results showed arachidonic acid metabolism was significantly different between the CP and HC groups. Glyceraldehyde-3-phosphate dehydrogenase, actin beta, phosphoglycerate mutase 1, Enolase 1, Myeloperoxidase, and actin alpha 1 were identified as potential proteins related to cholelithiasis. The correlation between the merged modules and clinical biochemical tests was calculated. A diagnostic panel composed of four candidate biomarkers, including 3-oxotetradecanoic acid, 12-hydroxydodecanoic acid, hemoglobin subunit delta (HBD), and fibrinogen beta chain (FGB), was proposed based on three modules that were significantly associated with cholelithiasis. This study provided some direct molecular insights into cholelithiasis by showing the differences in plasma metabolic and protein profiles between CPs and HCs and presented a potential biomarker panel with two metabolites (3-oxotetradecanoic acid, 12-hydroxydodecanoic acid) and two proteins (HBD, FGB) for predicting cholelithiasis. We also explored the potential correlation of clinical biochemical parameters with combined modules. These findings may provide some reference for the diagnosis of cholelithiasis in clinical practice.
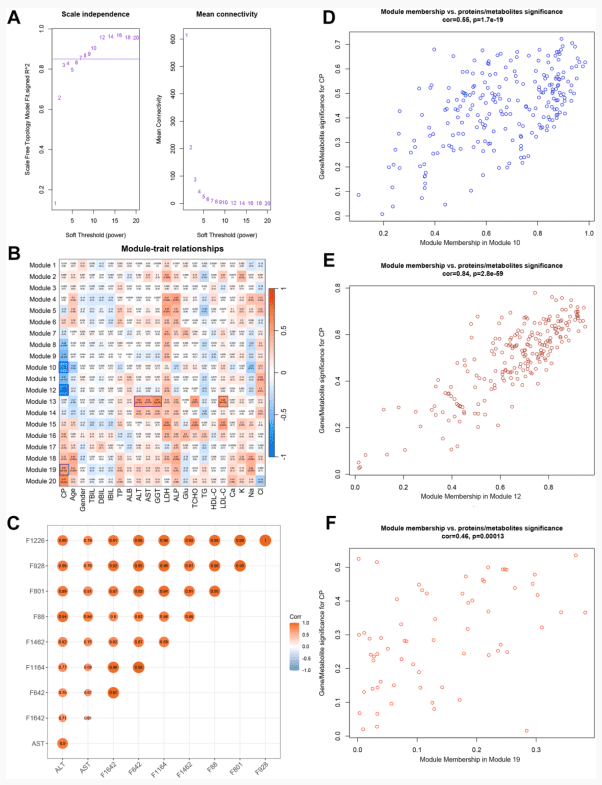 Integrome analysis of Proteomics and Metabolomics.
Integrome analysis of Proteomics and Metabolomics.
Next-Generation Omics Solutions:
Proteomics & Metabolomics
Ready to get started? Submit your inquiry or contact us at support-global@metwarebio.com.