Phosphoproteomics
Phosphoproteomics
Highly specific enrichment material Ti-IMAC for comprehensive coverage of modified proteins.
High-sensitivity and high-resolution mass spectrometry platform.
High-throughput 4D protein profiling detection technology.
Technology Introduction
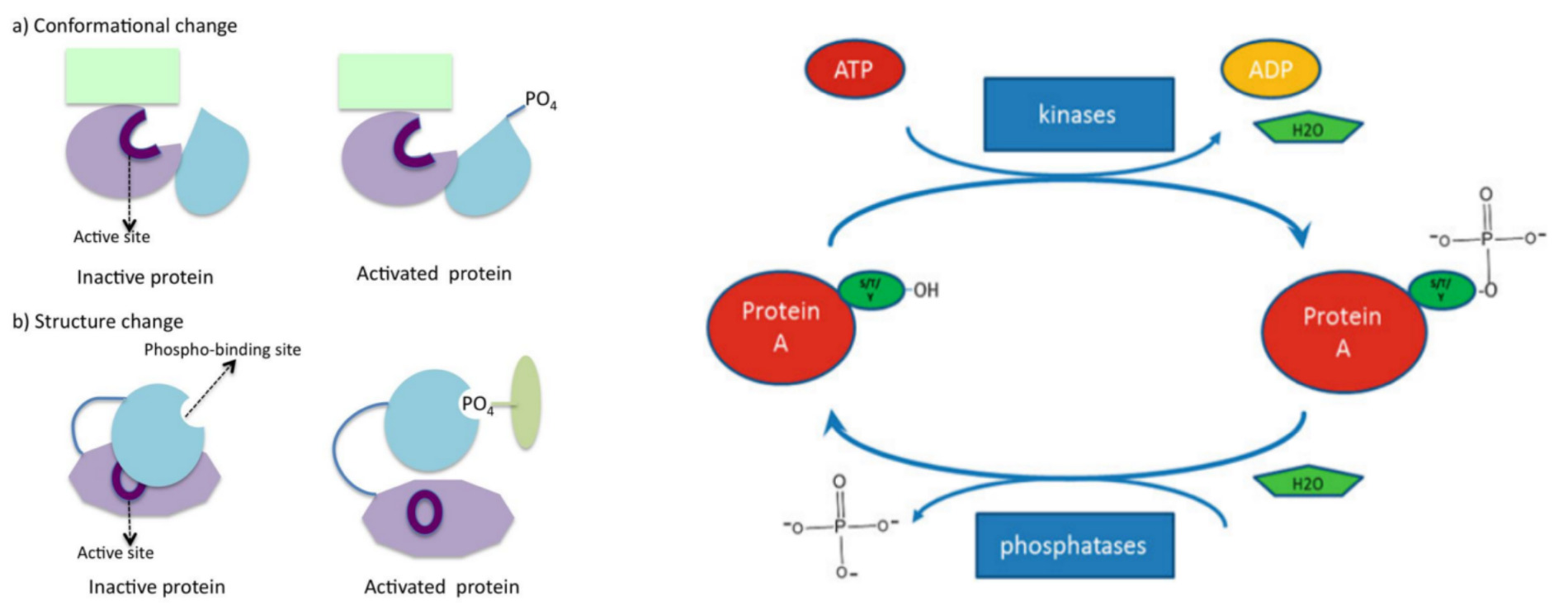
Protein phosphorylation refers to the process by which a protein transfers the phosphate group on ATP or GTP to a specific site (amino acid residue Ser, Tyr, Thr) on a protein under the catalysis of phosphorylase. Phosphorylation is a widely occurring post-translational modification, with over 30% of cellular proteins undergoing phosphorylation. Therefore, phosphorylation is the most fundamental, widespread, and important mechanism for regulating and controlling protein activity and function. Phosphorylation is involved in various physiological and pathological processes, regulating cellular proliferation, development, differentiation, apoptosis, and other life activities, and is widely applied in research fields such as signal transduction pathways, apoptosis, development and differentiation, and cancer mechanisms.
Phosphorylation proteomics aims to identifying phosphorylated proteins and peptides in diverse samples, accurately localizing and quantifying phosphorylation sites, and ultimately elucidating their biological functions. This methodology begins with protein sample digestion into peptide mixtures, followed by liquid chromatography to separate digested peptide components and reduce sample complexity. Subsequently, phosphorylation peptides are enriched using high-quality phosphorylation modification antibodies and biological materials. Finally, mass spectrometry technology with DDA (data-dependent acquisition) mode is employed for analysis and quantification.
Phosphorylation proteomics aims to identifying phosphorylated proteins and peptides in diverse samples, accurately localizing and quantifying phosphorylation sites, and ultimately elucidating their biological functions. This methodology begins with protein sample digestion into peptide mixtures, followed by liquid chromatography to separate digested peptide components and reduce sample complexity. Subsequently, phosphorylation peptides are enriched using high-quality phosphorylation modification antibodies and biological materials. Finally, mass spectrometry technology with DDA (data-dependent acquisition) mode is employed for analysis and quantification.
Applications of Phosphorylation Proteomics
Medical Research
Disease Mechanisms, Molecular Diagnosis, Biomarker Development, Drug Targets.
Animal Research
Reproductive Development, Disease Mechanisms, Nutrient Metabolism, Animal Toxicology.
Plant Research
Reproductive Development, Abiotic Stress Response, Disease Resistance, Crop Improvement.
Microbiology
Pathogenic Mechanisms, Drug Resistance, Stress-Related Proteins Screening, Environmental Impact Mechanisms.
SERVICES
Bile Acid
Oxylipin Targeted Metabolomics
Neurotransmitter Targeted Metabolomics
Steroid Hormone Targeted Metabolomics
Energy Metabolism
Tryptophan Targeted Metabolomics
Amino Acid Targeted Metabolomics
Short-Chain Fatty Acids
Plant Hormone Assay
Carotenoid Targeted Metabolomics
Anthocyanin Assay
Gibberellin Assay
Contact Us
Name can't be empty
Email error!
Message can't be empty
CONTACT FOR DEMO
Project Workflow of Phosphoproteomics

1
Biological Sample

2
Total Proteins

3
Total Peptides
4
Phosphopeptides

5
RAW Data

6
Bioinformation Report
Sample Requirements of Phosphoproteomics
| Sample Type | Samples | Recommended Sample Size | Minimum Sample Size |
| Human/Animal Tissue | Normal tissues (heart, liver, spleen, lungs, intestines, kidneys, etc.) | 50mg | 30mg |
| Fatty tissue | 1g | 500mg | |
| Brain tissue | 100mg | 50mg | |
| Bone | 1g | 500mg | |
| Hair | 1g | 500mg | |
| Plant Tissue | Young tissue (young leaf, seedling, petal, etc.) | 500mg | 200mg |
| Mature tissue (root, stem, fruit, pericarp, etc.) | 2g | 1.5g | |
| Liquid | Joint fluid, Lymph fluid | 1ml | 500μL |
| Cerebrospinal fluid | 1ml | 500μL | |
| Amniotic fluid | 5ml | 2ml | |
| Urine | 50mL | 20mL | |
| Fungi | 1g | 500mg | |
| Cells | Primary Cells | 2×10^7 | 1×10^7 |
| Transmissible cells | 1×10^7 | 5×10^6 | |
| Sperm, Platelets | 5×10^7 | 2×10^7 | |
| Protein | Protein | 1000μg | 500μg |
Biological duplicates: A minimum of 3 replicates is required; 3-6 replicates for animal samples; 6-10 for clinical samples.
WHAT'S NEXT IN OMICS: THE METABOLOME
Please submit a detailed description of your project. We will provide you with a customized project plan metabolomics services to meet your research requests. You can also send emails directly to support-global@metwarebio.com for inquiries.
Name can't be empty
Email error!
Message can't be empty
CONTACT FOR DEMO


