Metabolomics in Cancer Research and Emerging Applications
Cancer is a complex disease affected by genetic mutations and predisposition, environmental factors, and metabolic dysregulation. A lot of attention has been given to understanding the genomic aspect of cancer, especially in the last 10 years with the advance in sequencing technologies and the study of metabolomics in cancer research and emerging applications in clinical. A recent review by Dr. David Wishart (Metabolite 2022) described the multi-omics view of cancer and cancer metabolomics such as tumor metabolome and breast cancer metabolomics. In the david wishart metabolomics review, the genome, exposome, and metabolome all contribute to cancer development, and in turn, cancer cells further modify the genome, exposome, and metabolome through their altered metabolism and microenvironment (Figure 1).
With the emergence of high-throughput and high-resolution mass spectrometry, metabolomics is coming to the forefront as an essential aspect of cancer research. Many oncometabolites have been used as diagnostic biomarkers, including branched-chain amino acids and 2-hydroxy-glutarate. It has been estimated that hundreds of thousands of metabolites in humans are yet to be discovered and metabolomics is the prime technology for metabolite discovery and biomarker development.
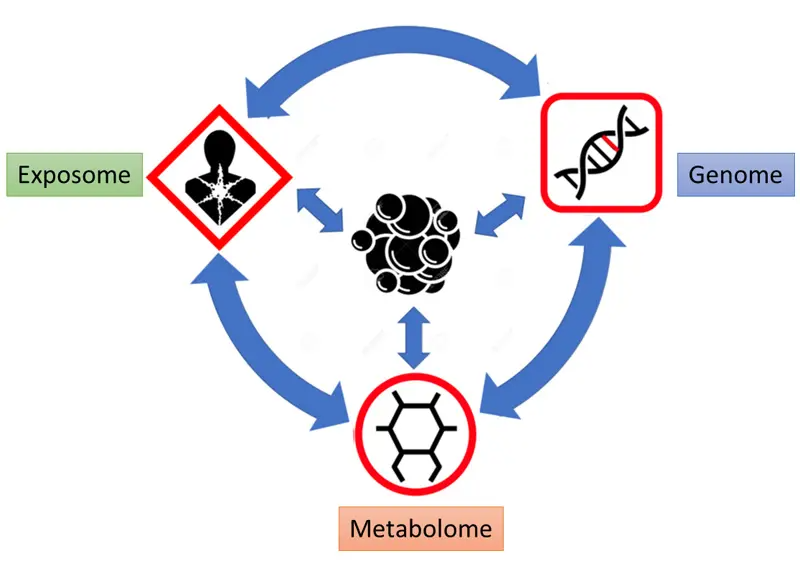
Figure 1. Multi-omics view of cancer development and subsequent feed-back. Figure adapted from Wishart D. Metabolomics and the Multi-Omics View of Cancer. Metabolite 2022, 12, 154.
Case Study: Precise pathological classification of non-small cell lung adenocarcinoma and squamous carcinoma based on an integrated platform of targeted metabolome and lipidome (Cao et al., Metabolomics 2021)
Lung adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC) are the most common subtypes of Non-small cell lung cancer. This study defines the metabolic differences between the two subtypes using LC-ESI-Q TRAP-MS/MS. A total of 128 plasma samples were tested and identified a total of 1141 compounds. Differential metabolites were screened based on Fold Change (FC) variations and VIP values (FC>2 or FC < 0.5 and VIP > 1.0). A total of 19 differential metabolites were identified, including 3 down-regulated and 16 up-regulated differential metabolites (Figure 2). Four of the differential metabolites (2-(Methylthio) ethanol, Cortisol, d-Glyceric Acid, and N-Acetylhistamine) were found to be diagnostic to separate LUAD and LUSC (Figure 3).
_1714291257_WNo_800d702.png)
Figure 2. The 19 differential metabolites between LUAD and LUSC
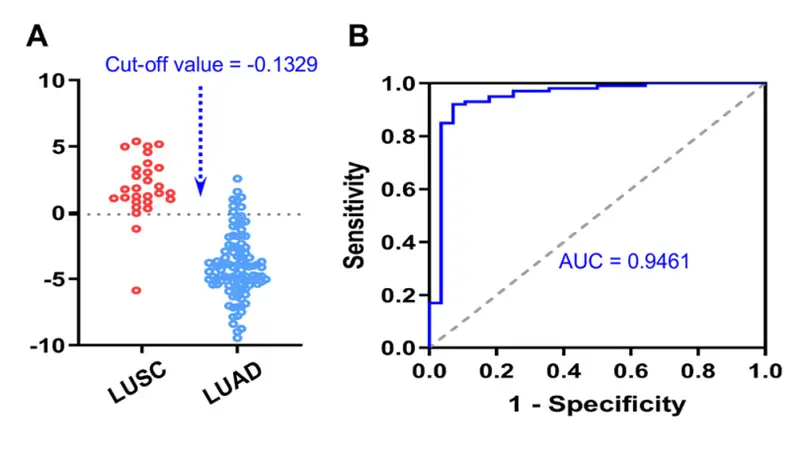
Figure 3. Combined diagnostic determination of 2-(Methylthio) ethanol, Cortisol, d-Glyceric Acid, and N-Acetylhistamine to distinguish LUAD and LUSC.
Bile Acid
Oxylipin Targeted Metabolomics
Neurotransmitter Targeted Metabolomics
Steroid Hormone Targeted Metabolomics
Energy Metabolism
Tryptophan Targeted Metabolomics
Amino Acid Targeted Metabolomics
Short-Chain Fatty Acids
Plant Hormone Assay
Carotenoid Targeted Metabolomics
Anthocyanin Assay
Gibberellin Assay
Contact Us
Name can't be empty
Email error!
Message can't be empty
CONTACT FOR DEMO


